

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0299.3
(762 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
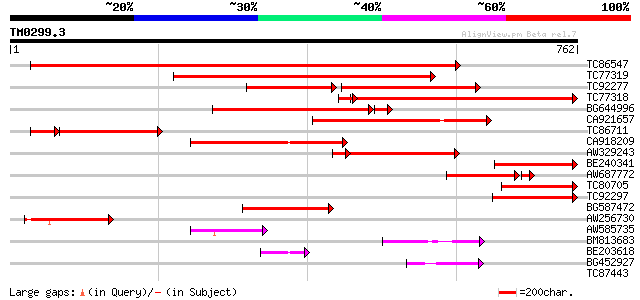
Score E
Sequences producing significant alignments: (bits) Value
TC86547 similar to GP|20805911|gb|AAM28891.1 NADPH oxidase {Nico... 657 0.0
TC77319 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 608 e-174
TC92277 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 298 e-143
TC77318 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 464 e-131
BG644996 similar to GP|20805911|gb NADPH oxidase {Nicotiana taba... 313 3e-90
CA921657 similar to PIR|T03826|T03 cytochrome b245 beta chain ho... 310 2e-84
TC86711 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 220 1e-64
CA918209 weakly similar to GP|16549089|db respiratory burst oxid... 243 2e-64
AW329243 similar to GP|16549089|db respiratory burst oxidase hom... 229 4e-60
BE240341 similar to GP|16549089|db respiratory burst oxidase hom... 194 1e-49
AW687772 similar to GP|16549089|db respiratory burst oxidase hom... 166 7e-46
TC80705 similar to GP|19715905|emb|CAC84140. NADPH oxidase {Nico... 148 7e-36
TC92297 weakly similar to GP|19715905|emb|CAC84140. NADPH oxidas... 130 2e-30
BG587472 similar to GP|20160800|db putative respiratory burst ox... 115 8e-26
AW256730 weakly similar to GP|9757922|dbj respiratory burst oxid... 93 3e-19
AW585735 similar to GP|13958313|gb| NADPH oxidase 1 {Podospora a... 63 4e-10
BM813683 weakly similar to GP|15341529|gb ferric-chelate reducta... 61 1e-09
BE203618 weakly similar to GP|3242787|gb|A respiratory burst oxi... 48 1e-05
BG452927 weakly similar to GP|20268772|gb putative FRO2; NADPH o... 45 8e-05
TC87443 similar to GP|13397927|emb|CAC34625. putative calmodulin... 41 0.002
>TC86547 similar to GP|20805911|gb|AAM28891.1 NADPH oxidase {Nicotiana
tabacum}, partial (57%)
Length = 2214
Score = 657 bits (1694), Expect = 0.0
Identities = 335/585 (57%), Positives = 423/585 (72%), Gaps = 7/585 (1%)
Frame = +1
Query: 29 KVDRYKSGATHALQCFRVMTKNVVFEGWSEVEKRFDGLAI--EGKLLKTRFNECIGMSE- 85
+++R KS HAL + ++K V GWSEVEK FD L + +G L +T F +CIG++E
Sbjct: 433 RLERTKSAVAHALTGLKFISKTDVGAGWSEVEKVFDKLTVTTDGYLPRTLFAKCIGLNEE 612
Query: 86 LKDFACKLFDALARCRGITSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRIN 145
K +A LFD LAR RGI SI K + RE W+ I++QSFD+R++ FFD+VDK+ADGRI
Sbjct: 613 SKAYAEMLFDTLARQRGIQGGSINKIQFREFWDCISDQSFDTRLKIFFDMVDKDADGRIT 792
Query: 146 EEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQASAQST 205
EE++K II LSA++NKLS IQ+++ EYA+LIMEELDPD+ GYI + +LE LLL ++T
Sbjct: 793 EEEIKNIICLSATANKLSNIQKQAEEYAALIMEELDPDDTGYILIGNLETLLLHGPEETT 972
Query: 206 T-DSKVVSQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFTWKF 264
+SK +SQMLSQK PT E NPIKR + YF QDNW+R WV ALW+ + + LF +KF
Sbjct: 973 RGESKYLSQMLSQKLRPTFEGNPIKRWYRDTKYFFQDNWRRSWVFALWIGVMLGLFAFKF 1152
Query: 265 FQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPFDDN 324
QY+ R+ + VMG+CV +AKG AETLK NMA+IL P+CRNTITWLR+KTKLG VPFDD
Sbjct: 1153 VQYRRRSAYKVMGHCVCMAKGAAETLKLNMAIILLPVCRNTITWLRNKTKLGAFVPFDDG 1332
Query: 325 VNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLKGT 384
+NFHK+IA IAIG+G+HA+ HL DFPR+LH + +Y+ ++PFFG+ KP NYW F+K
Sbjct: 1333 LNFHKMIALAIAIGVGIHAIYHLA*DFPRILHASNEKYKLIEPFFGE-KPTNYWHFVKSW 1509
Query: 385 EGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIH 444
EG TG+LMVVLMAI F LA RRNR KLPKP KLTGFNAFWYSHHL IIVY +L+IH
Sbjct: 1510 EGVTGILMVVLMAIAFTLANTRFRRNRTKLPKPFNKLTGFNAFWYSHHLFIIVYAMLIIH 1689
Query: 445 GHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKMS 504
G LYL+K+W TTWMYLA+P+ +Y ERLIRA RS K VRI V+ Y GNV A+ MS
Sbjct: 1690 GTKLYLTKEWNHKTTWMYLAIPITIYGLERLIRALRSSIKSVRILKVAVYPGNVLAINMS 1869
Query: 505 KPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKA 564
KPQGF Y SGQY+ VNC+ +SP EWHPFSITSAP DDYLSVHI+ LGDWT L+
Sbjct: 1870 KPQGFSYKSGQYMLVNCAAVSPLEWHPFSITSAPNDDYLSVHIKILGDWTRSLKTKFLTG 2049
Query: 565 CQPSSD-DQKCILQANTLPD-TSKPRIPRLWIDGP-YGSPAQDYK 606
S + + N + T++ P++ IDGP + PAQDY+
Sbjct: 2050 LPASHQWTEWTF*ELNA*KEITAQVLFPKVLIDGPVWERPAQDYR 2184
>TC77319 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (42%)
Length = 1137
Score = 608 bits (1568), Expect = e-174
Identities = 275/352 (78%), Positives = 313/352 (88%)
Frame = +2
Query: 221 PTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFTWKFFQYKNRAVFDVMGYCV 280
PTKE NPIKR L++L YFI+DNWKRIWV+ALWLSIC ALFTWKF QYKNR+ F VMGYCV
Sbjct: 2 PTKEYNPIKRSLRSLNYFIEDNWKRIWVIALWLSICAALFTWKFIQYKNRSAFRVMGYCV 181
Query: 281 VIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPFDDNVNFHKVIAFGIAIGIG 340
IAKG AETLKFNMALIL P+CRNTITWLRSKTKLG+VVPFDDN+NFHKVIAFGIAIG+G
Sbjct: 182 TIAKGAAETLKFNMALILMPVCRNTITWLRSKTKLGVVVPFDDNINFHKVIAFGIAIGVG 361
Query: 341 LHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLKGTEGWTGVLMVVLMAIPF 400
LHA++HLTCDFPRLLH T AEY P+K FFG ++P+NYWWF+KGTEGWTG++MVVLMAI F
Sbjct: 362 LHAISHLTCDFPRLLHATDAEYVPMKKFFGDHRPNNYWWFVKGTEGWTGIVMVVLMAIAF 541
Query: 401 ILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHGHFLYLSKKWYKNTTW 460
LAQPW RRN+LKLP LKKLTGFNAFWYSHHL +IVY LL++HG+FLYLSKKWYK TTW
Sbjct: 542 TLAQPWFRRNKLKLPPLLKKLTGFNAFWYSHHLFVIVYALLIVHGYFLYLSKKWYKKTTW 721
Query: 461 MYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVN 520
MYLA+PMI+Y ERL+RAFRS +K V+I V+ Y GNV AL +SKPQGFKY SGQYI+VN
Sbjct: 722 MYLAIPMIIYACERLLRAFRSGYKSVKILKVAVYPGNVLALHVSKPQGFKYHSGQYIFVN 901
Query: 521 CSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQ 572
C+D+SPF+WHPFSITSAP DDY+SVHIRT GDWTSQL+A+ KACQP+S DQ
Sbjct: 902 CADVSPFQWHPFSITSAPGDDYVSVHIRTAGDWTSQLKAVFAKACQPASGDQ 1057
>TC92277 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (34%)
Length = 949
Score = 298 bits (764), Expect(2) = e-143
Identities = 147/187 (78%), Positives = 161/187 (85%), Gaps = 1/187 (0%)
Frame = +3
Query: 447 FLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSKP 506
FLYLSKKWYK TTWMYLAVPMILY ERL+ AFRS +K VRI V+ Y GNV AL SKP
Sbjct: 387 FLYLSKKWYKKTTWMYLAVPMILYGCERLLPAFRSGYKSVRILKVAVYPGNVLALHASKP 566
Query: 507 QGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKACQ 566
QGFKY SGQYIYVNCSDISPFEWHPFSITSAP DDY+SVHIRTLGDWTSQL+ I KACQ
Sbjct: 567 QGFKYTSGQYIYVNCSDISPFEWHPFSITSAPGDDYISVHIRTLGDWTSQLKGIFAKACQ 746
Query: 567 PSSDDQKCILQANTLP-DTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLIS 625
P++DDQ +L+A+ LP +S PR+PRL IDGPYG+PAQDYKNYEVLLLVGLGIGATPLIS
Sbjct: 747 PANDDQSGLLRADMLPGKSSLPRMPRLRIDGPYGAPAQDYKNYEVLLLVGLGIGATPLIS 926
Query: 626 ILKGMLN 632
ILK +LN
Sbjct: 927 ILKDVLN 947
Score = 230 bits (586), Expect(2) = e-143
Identities = 101/121 (83%), Positives = 111/121 (91%)
Frame = +1
Query: 319 VPFDDNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYW 378
VPFDDN+NFHKVIAFGIAIG+GLHA++HLTCDFPRLLH T EYEP+K FFG +P+NYW
Sbjct: 1 VPFDDNINFHKVIAFGIAIGVGLHAISHLTCDFPRLLHATDEEYEPMKQFFGDERPNNYW 180
Query: 379 WFLKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVY 438
WF+KGTEGWTGV+MVVLMAI FILAQPW RRNRL LPKPLKKLTGFNAFWYSHHL +IVY
Sbjct: 181 WFVKGTEGWTGVVMVVLMAIAFILAQPWFRRNRLNLPKPLKKLTGFNAFWYSHHLFVIVY 360
Query: 439 V 439
V
Sbjct: 361 V 363
>TC77318 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (38%)
Length = 1394
Score = 464 bits (1195), Expect = e-131
Identities = 218/305 (71%), Positives = 262/305 (85%)
Frame = +2
Query: 458 TTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYI 517
TTWMYLA+PMI+Y ERL+RAFRS +K V+I V+ Y GNV AL +SKPQGFKY SGQYI
Sbjct: 83 TTWMYLAIPMIIYACERLLRAFRSGYKSVKILKVAVYPGNVLALHVSKPQGFKYHSGQYI 262
Query: 518 YVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQ 577
+VNC+D+SPF+WHPFSITSAP DDY+SVHIRT GDWTSQL+A+ KACQP+S DQ +L+
Sbjct: 263 FVNCADVSPFQWHPFSITSAPGDDYVSVHIRTAGDWTSQLKAVFAKACQPASGDQSGLLR 442
Query: 578 ANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQ 637
A+ L + PR+P+L IDGPYG+PAQDYK+YEV+LLVGLGIGATPLISILK +LNN+KQQ
Sbjct: 443 ADMLQGNNIPRMPKLLIDGPYGAPAQDYKDYEVILLVGLGIGATPLISILKDVLNNMKQQ 622
Query: 638 QDLENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHN 697
+D+E GV+ES VK NK+K FAT +AYFYWVTREQGSFEWFKG+M+EIA+ DK+G+IELHN
Sbjct: 623 KDIEQGVVESGVKNNKRKPFATNRAYFYWVTREQGSFEWFKGVMDEIADYDKDGLIELHN 802
Query: 698 YCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDK 757
YCT VY+EGD SALITMLQSL+ +K G+D VS TRVKTHF RPNWR V++H +LKHP K
Sbjct: 803 YCTSVYEEGDARSALITMLQSLHHAKSGVDIVSGTRVKTHFARPNWRTVFKHTALKHPGK 982
Query: 758 RVGKY 762
RVG +
Sbjct: 983 RVGVF 997
Score = 50.8 bits (120), Expect = 2e-06
Identities = 18/26 (69%), Positives = 23/26 (88%)
Frame = -2
Query: 442 VIHGHFLYLSKKWYKNTTWMYLAVPM 467
++HG+FLYLSKKWYK TTWMY+ + M
Sbjct: 79 IVHGYFLYLSKKWYKKTTWMYIIIGM 2
>BG644996 similar to GP|20805911|gb NADPH oxidase {Nicotiana tabacum},
partial (25%)
Length = 726
Score = 313 bits (802), Expect(2) = 3e-90
Identities = 141/216 (65%), Positives = 175/216 (80%)
Frame = +3
Query: 273 FDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPFDDNVNFHKVIA 332
++VMG+CV +AKG AETLK NMA+IL P+CRNTITWLR+KTKLG+ VPFDDN+NFHKVIA
Sbjct: 3 YEVMGHCVCMAKGAAETLKLNMAIILLPVCRNTITWLRNKTKLGIAVPFDDNLNFHKVIA 182
Query: 333 FGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLKGTEGWTGVLM 392
+A G+G+HA+ HLTCDFPRLLH + +Y+ ++PFFGK +P +YW F+K EG TG++M
Sbjct: 183 VAVATGVGIHAIYHLTCDFPRLLHANSEKYKLMEPFFGK-QPTSYWHFVKSWEGVTGIVM 359
Query: 393 VVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHGHFLYLSK 452
VVLM I F LA PW R+ R+KLPKPL LTGFNAFWYSHHL + VY LLV+HG LYL++
Sbjct: 360 VVLMTIAFTLASPWFRKGRVKLPKPLNSLTGFNAFWYSHHLFVFVYALLVVHGIKLYLTR 539
Query: 453 KWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRI 488
+W+K TTWMYL +P+I+Y ERL RA RS K VRI
Sbjct: 540 EWHKKTTWMYLVIPIIIYALERLTRALRSSIKPVRI 647
Score = 38.1 bits (87), Expect(2) = 3e-90
Identities = 17/24 (70%), Positives = 19/24 (78%)
Frame = +1
Query: 491 VSWYTGNVFALKMSKPQGFKYMSG 514
V+ Y GNV AL MSKPQGF+Y SG
Sbjct: 655 VAVYPGNVLALHMSKPQGFRYKSG 726
>CA921657 similar to PIR|T03826|T03 cytochrome b245 beta chain homolog
RbohAp108 - Arabidopsis thaliana, partial (26%)
Length = 749
Score = 310 bits (793), Expect = 2e-84
Identities = 145/240 (60%), Positives = 184/240 (76%)
Frame = -2
Query: 408 RRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPM 467
RRN +KLP+P +LTGFNAFWYSHHL +IVYVLL+IHG LYL KWY TTWMYL VP+
Sbjct: 748 RRNLIKLPEPFSRLTGFNAFWYSHHLFVIVYVLLIIHGVKLYLVHKWYYQTTWMYLTVPL 569
Query: 468 ILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPF 527
+LY SER +R FRS VR+ V+ Y GNV L+MSKP F+Y SGQY++V C+ +SPF
Sbjct: 568 LLYASERTLRLFRSGLYTVRLIKVAIYPGNVLTLQMSKPPQFRYKSGQYMFVQCAAVSPF 389
Query: 528 EWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKP 587
EWHPFSITSAP DDYLSVHIR LGDWT +L+ + + C+P + +L+A+ +T+K
Sbjct: 388 EWHPFSITSAPGDDYLSVHIRQLGDWTQELKRVFSEVCEPPVSGRSGLLRAD---ETTKK 218
Query: 588 RIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQQDLENGVLES 647
+P+L IDGPYG+PAQDY+ Y+VLLLVGLGIGATP ISILK +LNNI + ++L + V ++
Sbjct: 217 SLPKLKIDGPYGAPAQDYRKYDVLLLVGLGIGATPFISILKDLLNNIVKMEELADSVSDT 38
>TC86711 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (27%)
Length = 1075
Score = 220 bits (560), Expect(2) = 1e-64
Identities = 109/138 (78%), Positives = 130/138 (93%)
Frame = +2
Query: 68 IEGKLLKTRFNECIGMSELKDFACKLFDALARCRGITSTSITKDELREIWEQITNQSFDS 127
++GKL KTRF++CIGM+E KDFA +LFDALAR RGITS SITKDELR+ WEQIT+QSFDS
Sbjct: 653 VDGKLPKTRFSQCIGMNESKDFAGELFDALARRRGITSASITKDELRQFWEQITDQSFDS 832
Query: 128 RVQTFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGY 187
R+QTFFD+VDKNADGRI+E++VKEII+LSAS+NKLSK+QER+ EYA+LIMEELDPDNLG+
Sbjct: 833 RLQTFFDMVDKNADGRISEDEVKEIITLSASANKLSKLQERAEEYAALIMEELDPDNLGF 1012
Query: 188 IELYSLEMLLLQASAQST 205
IEL++LEMLLLQA AQST
Sbjct: 1013IELHNLEMLLLQAPAQST 1066
Score = 45.8 bits (107), Expect(2) = 1e-64
Identities = 24/39 (61%), Positives = 28/39 (71%), Gaps = 1/39 (2%)
Frame = +1
Query: 29 KVDRYKSGATHALQCFRVMTKNV-VFEGWSEVEKRFDGL 66
+VDR KSGA AL+ + MTKNV GWS+VEKRFD L
Sbjct: 532 RVDRAKSGAARALKGLKFMTKNVGTDRGWSQVEKRFDEL 648
>CA918209 weakly similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (20%)
Length = 713
Score = 243 bits (621), Expect = 2e-64
Identities = 114/213 (53%), Positives = 150/213 (69%), Gaps = 1/213 (0%)
Frame = +3
Query: 243 WKRIWVMALWLSICVALFTWKFFQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPIC 302
W+R WV+ LWL C LF WKF QY R+ F+VMGYC+ AKG AETLK NMALIL P+C
Sbjct: 72 WRRSWVVVLWLVGCFGLFGWKFDQYSKRSGFEVMGYCLPTAKGAAETLKLNMALILLPVC 251
Query: 303 RNTITWLRSKTKLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEY 362
RNTITWLR+ +L V+PF+DN+NFHK+IA GI +G+ LH HLTCDFPR+ + + +
Sbjct: 252 RNTITWLRNDPRLNYVIPFNDNINFHKLIAGGIVVGVILHGGTHLTCDFPRISDSDKSIF 431
Query: 363 -EPLKPFFGKNKPDNYWWFLKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKL 421
+ + FG ++P Y L TE +G+ MV++MAI F LA W RR LP ++++
Sbjct: 432 RQTIAAGFGYHQP-TYMEILATTEVASGIGMVLIMAIAFSLATKWPRRRSPVLPLSIRRV 608
Query: 422 TGFNAFWYSHHLLIIVYVLLVIHGHFLYLSKKW 454
TG+N FWYSHHL +IVYVLL++H FL+L+ KW
Sbjct: 609 TGYNTFWYSHHLFVIVYVLLIVHSMFLFLADKW 707
>AW329243 similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (18%)
Length = 627
Score = 229 bits (583), Expect = 4e-60
Identities = 118/171 (69%), Positives = 134/171 (78%), Gaps = 1/171 (0%)
Frame = +3
Query: 435 IIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWY 494
I ++LL+I F Y K TWMYLAVPMILY ERL+RAFRS +K VRI V+ Y
Sbjct: 120 ICFFILLIIW*IFCY--KIM*HI*TWMYLAVPMILYGCERLLRAFRSGYKSVRILKVAVY 293
Query: 495 TGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWT 554
GNV AL SKPQGFKY SGQYIYVNCSDISPFEWHPFSITSAP DDY+SVHIRTLGDWT
Sbjct: 294 PGNVLALHASKPQGFKYTSGQYIYVNCSDISPFEWHPFSITSAPGDDYISVHIRTLGDWT 473
Query: 555 SQLQAILEKACQPSSDDQKCILQANTLP-DTSKPRIPRLWIDGPYGSPAQD 604
SQL+ I KACQP++DDQ +L+A+ LP +S PR+PRL IDGPYG+PAQD
Sbjct: 474 SQLKGIFAKACQPANDDQSGLLRADMLPGKSSLPRMPRLRIDGPYGAPAQD 626
Score = 47.8 bits (112), Expect = 2e-05
Identities = 19/24 (79%), Positives = 22/24 (91%)
Frame = +2
Query: 435 IIVYVLLVIHGHFLYLSKKWYKNT 458
+IVYVL +IHG+FLYLSKKWYK T
Sbjct: 5 VIVYVLFIIHGYFLYLSKKWYKKT 76
>BE240341 similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (16%)
Length = 569
Score = 194 bits (493), Expect = 1e-49
Identities = 88/111 (79%), Positives = 101/111 (90%)
Frame = +1
Query: 652 NKKKHFATKQAYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHNYCTGVYKEGDDGSA 711
NKK+ FATK+AYFYWVTREQGSFEW KG+MNE+AENDKEGVIELHNYCT VY+EGD SA
Sbjct: 4 NKKRPFATKRAYFYWVTREQGSFEWLKGVMNEVAENDKEGVIELHNYCTSVYEEGDARSA 183
Query: 712 LITMLQSLYQSKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDKRVGKY 762
LITMLQSL+ +K G+D VSETRVKTHF RPNWRNVY+HV+LKHP+K+VG +
Sbjct: 184 LITMLQSLHHAKNGVDVVSETRVKTHFARPNWRNVYKHVALKHPEKKVGVF 336
>AW687772 similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (13%)
Length = 622
Score = 166 bits (419), Expect(2) = 7e-46
Identities = 77/98 (78%), Positives = 91/98 (92%)
Frame = +1
Query: 588 RIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQQDLENGVLES 647
R+P+L IDGPYG+PAQDYK+YEV+LLVGLGIGATPLISILK +LNN+KQQ+D+E GV+ES
Sbjct: 232 RMPKLLIDGPYGAPAQDYKDYEVILLVGLGIGATPLISILKDVLNNMKQQKDIEQGVVES 411
Query: 648 RVKTNKKKHFATKQAYFYWVTREQGSFEWFKGLMNEIA 685
VK NK+K FAT +AYFYWVTREQGSFEWFKG+M+EIA
Sbjct: 412 GVKNNKRKPFATNRAYFYWVTREQGSFEWFKGVMDEIA 525
Score = 37.4 bits (85), Expect(2) = 7e-46
Identities = 14/18 (77%), Positives = 17/18 (93%)
Frame = +3
Query: 688 DKEGVIELHNYCTGVYKE 705
DK+G+IELHNYCT VY+E
Sbjct: 531 DKDGLIELHNYCTSVYEE 584
>TC80705 similar to GP|19715905|emb|CAC84140. NADPH oxidase {Nicotiana
tabacum}, partial (14%)
Length = 837
Score = 148 bits (374), Expect = 7e-36
Identities = 62/101 (61%), Positives = 84/101 (82%)
Frame = +1
Query: 662 AYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQ 721
AYFYWVTREQGSF+WFKG+MN++AE D+ G+IELH+YCT VY++GD SALI M+QS+
Sbjct: 1 AYFYWVTREQGSFDWFKGVMNDVAEEDRRGLIELHSYCTSVYEQGDARSALIAMVQSINH 180
Query: 722 SKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDKRVGKY 762
+K+G+D VS TRV +HF +PNWR VY+ ++L HP+ +VG +
Sbjct: 181 AKHGVDVVSGTRVMSHFAKPNWRTVYKRIALNHPEAQVGVF 303
>TC92297 weakly similar to GP|19715905|emb|CAC84140. NADPH oxidase
{Nicotiana tabacum}, partial (14%)
Length = 719
Score = 130 bits (327), Expect = 2e-30
Identities = 56/113 (49%), Positives = 82/113 (72%)
Frame = +3
Query: 650 KTNKKKHFATKQAYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHNYCTGVYKEGDDG 709
K +F T++AYFYW+ EQG F+WFKG++NE+AE D + VIE+H++ T VY +GD
Sbjct: 6 KKTDLSNFKTRKAYFYWMAGEQGFFDWFKGVINEVAEEDHKKVIEIHSHLTSVYXDGDAR 185
Query: 710 SALITMLQSLYQSKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDKRVGKY 762
SAL+T+LQSL +K GLD ++ T + ++F RPNWR+VY ++ HP KR+G +
Sbjct: 186 SALVTVLQSLNHAKEGLDILNGTPIASYFARPNWRSVYNRIANNHPQKRIGVF 344
>BG587472 similar to GP|20160800|db putative respiratory burst oxidase
protein {Oryza sativa (japonica cultivar-group)},
partial (13%)
Length = 387
Score = 115 bits (287), Expect = 8e-26
Identities = 53/123 (43%), Positives = 76/123 (61%)
Frame = +3
Query: 313 TKLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKN 372
T L ++PFDDN+NFHK+IA G+ IG +H H++CDFPRL+ ++ +
Sbjct: 18 TFLSQIIPFDDNINFHKIIAVGVVIGTLIHVGVHVSCDFPRLVSCPTEKFMAILGSGFDY 197
Query: 373 KPDNYWWFLKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHH 432
K +Y + G TG+ MV++MA F LA + R++ + LP PL L GFN+FWY+HH
Sbjct: 198 KQPSYLTLVTSPPGITGIFMVLIMAFSFTLATHYFRKSVVTLPSPLHHLAGFNSFWYAHH 377
Query: 433 LLI 435
LLI
Sbjct: 378 LLI 386
>AW256730 weakly similar to GP|9757922|dbj respiratory burst oxidase protein
{Arabidopsis thaliana}, partial (11%)
Length = 681
Score = 93.2 bits (230), Expect = 3e-19
Identities = 52/125 (41%), Positives = 78/125 (61%), Gaps = 5/125 (4%)
Frame = +2
Query: 20 NSNGGCDKLKVDRYKSGATHALQCFRVMTKNVV---FEGWSEVEKRFDGLAIEGKLLKTR 76
N+NG K+ R +SGA L+ RV+ + V + W +EKRF A++G L K +
Sbjct: 317 NNNGS----KMRRMESGAARGLKGLRVLDRTVTGKEADAWKSIEKRFTQHAVDGMLSKDK 484
Query: 77 FNECIGM-SELKDFACKLFDALARCRGITSTS-ITKDELREIWEQITNQSFDSRVQTFFD 134
F C+GM ++ KDFA +L++ALAR R I + + IT +E R WE +TN+ +S++Q FFD
Sbjct: 485 FGTCMGMGADSKDFAGELYEALARRRNICAENGITLNEARVFWEDMTNKDLESKLQVFFD 664
Query: 135 LVDKN 139
+ DKN
Sbjct: 665 MCDKN 679
>AW585735 similar to GP|13958313|gb| NADPH oxidase 1 {Podospora anserina},
partial (4%)
Length = 496
Score = 63.2 bits (152), Expect = 4e-10
Identities = 35/106 (33%), Positives = 54/106 (50%), Gaps = 3/106 (2%)
Frame = +1
Query: 244 KRIWVMALWLSICVALFTWKFFQYKNRAVFD---VMGYCVVIAKGGAETLKFNMALILFP 300
KR+ W + LF + F + K D V+G V ++G L ++ ALIL P
Sbjct: 103 KRLAFWIFWFGSHIGLFIYGFLKQKTDHELDNLNVIGLSVWSSRGAGLCLAYDGALILLP 282
Query: 301 ICRNTITWLRSKTKLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAH 346
+CRN I RS L ++PFD+N+ FH+ A+ + I +H +H
Sbjct: 283 VCRNLIKQFRSFRFLNNIIPFDENLWFHRQTAYAMLIFTLIHTFSH 420
>BM813683 weakly similar to GP|15341529|gb ferric-chelate reductase {Pisum
sativum}, partial (21%)
Length = 523
Score = 61.2 bits (147), Expect = 1e-09
Identities = 38/140 (27%), Positives = 67/140 (47%), Gaps = 2/140 (1%)
Frame = +1
Query: 501 LKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAP--EDDYLSVHIRTLGDWTSQLQ 558
L SK Y +++N +S +WHPF+++S+ E + LSV I+ +G W+++L
Sbjct: 25 LNFSKDPSLYYNPTSLVFINVPKVSKLQWHPFTVSSSCNLETNCLSVTIKNVGSWSNKLY 204
Query: 559 AILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGI 618
L SS D + ++GPYG + + +E + +V G
Sbjct: 205 QELSS----SSLDHLNVS-----------------VEGPYGPHSAQFLRHEQIAMVSGGS 321
Query: 619 GATPLISILKGMLNNIKQQQ 638
G TP ISI++ ++ +QQ+
Sbjct: 322 GITPFISIIRDLIFQSQQQE 381
>BE203618 weakly similar to GP|3242787|gb|A respiratory burst oxidase protein
E {Arabidopsis thaliana}, partial (6%)
Length = 206
Score = 48.1 bits (113), Expect = 1e-05
Identities = 25/67 (37%), Positives = 38/67 (56%)
Frame = +3
Query: 337 IGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLKGTEGWTGVLMVVLM 396
+GI +HA H++CDFP L++++ ++ + F KP Y EG TG+ M+ LM
Sbjct: 6 VGILVHAGNHISCDFPLLVNSSPEKFSLIASDFNNKKP-TYKSLFISIEGVTGITMLTLM 182
Query: 397 AIPFILA 403
I F LA
Sbjct: 183 VISFTLA 203
>BG452927 weakly similar to GP|20268772|gb putative FRO2; NADPH oxidase
{Arabidopsis thaliana}, partial (17%)
Length = 610
Score = 45.4 bits (106), Expect = 8e-05
Identities = 30/107 (28%), Positives = 54/107 (50%), Gaps = 4/107 (3%)
Frame = +1
Query: 534 ITSAPED--DYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKP--RI 589
++S+P D ++++V I+ LG WT L+ + I + D S P +
Sbjct: 1 VSSSPLDGKNHIAVLIKVLGKWTGGLR--------------ERITDGDATEDLSVPPHMV 138
Query: 590 PRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQ 636
++GPYG + YE L+LV GIG +P ++IL +L+ +++
Sbjct: 139 VTASVEGPYGHEVPYHLMYENLILVAGGIGLSPFLAILSDILHRVRE 279
>TC87443 similar to GP|13397927|emb|CAC34625. putative calmodulin-related
protein {Medicago sativa}, partial (69%)
Length = 918
Score = 40.8 bits (94), Expect = 0.002
Identities = 22/71 (30%), Positives = 38/71 (52%)
Frame = +3
Query: 120 ITNQSFDSRVQTFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEYASLIMEE 179
++ + D V+ F DKN DG+I+ ++KE++ L+ E + E +MEE
Sbjct: 3 LSPSAMDDEVRKIFSKFDKNGDGKISRSELKEML--------LTLGSETTSEEVKRMMEE 158
Query: 180 LDPDNLGYIEL 190
LD + G+I+L
Sbjct: 159 LDQNGDGFIDL 191
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,845,358
Number of Sequences: 36976
Number of extensions: 413886
Number of successful extensions: 2157
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 2120
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2141
length of query: 762
length of database: 9,014,727
effective HSP length: 104
effective length of query: 658
effective length of database: 5,169,223
effective search space: 3401348734
effective search space used: 3401348734
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0299.3


