

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0299.13
(259 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
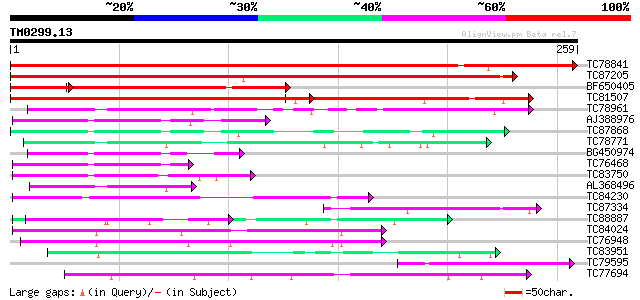
Score E
Sequences producing significant alignments: (bits) Value
TC78841 similar to GP|5815235|gb|AAD52609.1| splicing factor SR1... 450 e-127
TC87205 similar to GP|12324445|gb|AAG52185.1 putative splicing f... 294 2e-80
BF650405 similar to GP|20161426|db putative pre-mRNA splicing fa... 180 6e-57
TC81507 similar to GP|22831146|dbj|BAC16007. putative pre-mRNA s... 206 7e-54
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 86 2e-17
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 67 9e-12
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 67 9e-12
TC78771 similar to GP|16612268|gb|AAL27502.1 AT3g61860/F21F14_30... 65 3e-11
BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR p... 62 2e-10
TC76468 similar to PIR|E84638|E84638 probable RSZp22 splicing fa... 62 3e-10
TC83750 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 61 5e-10
AL368496 similar to GP|16612268|gb| AT3g61860/F21F14_30 {Arabido... 58 3e-09
TC84230 54 5e-08
TC87334 similar to GP|15293081|gb|AAK93651.1 unknown protein {Ar... 53 1e-07
TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 51 4e-07
TC84024 similar to PIR|S77714|S77714 RNA-binding protein precurs... 51 5e-07
TC76948 similar to SP|Q08935|ROC1_NICSY 29 kDa ribonucleoprotein... 50 7e-07
TC83951 similar to GP|22136316|gb|AAM91236.1 putative RNA bindin... 49 1e-06
TC79595 similar to GP|3850823|emb|CAA77136.1 U2 snRNP auxiliary ... 49 1e-06
TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-... 48 3e-06
>TC78841 similar to GP|5815235|gb|AAD52609.1| splicing factor SR1
{Arabidopsis thaliana}, partial (74%)
Length = 1380
Score = 450 bits (1157), Expect = e-127
Identities = 225/261 (86%), Positives = 239/261 (91%), Gaps = 2/261 (0%)
Frame = +3
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAED 60
MS RSSRT+YVGNLPGDIRLREVED+FYK+GPIVDIDLKIPP+PPGYAFV+FEDARDA+D
Sbjct: 42 MSGRSSRTLYVGNLPGDIRLREVEDLFYKYGPIVDIDLKIPPKPPGYAFVEFEDARDAQD 221
Query: 61 AIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPP 120
AIYYRDGYDFDG+RL VELAHGGRGSSSSVDRYS +SG S SRG SRRSDYRVLVTGLPP
Sbjct: 222 AIYYRDGYDFDGYRLLVELAHGGRGSSSSVDRYSRHSGRSGSRGVSRRSDYRVLVTGLPP 401
Query: 121 SASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRA 180
SASWQDLKDHMRKAG VCFSQVFRERGG+TGIVDYTNYDD+KYA+RKLDDSEFRNAFSR+
Sbjct: 402 SASWQDLKDHMRKAGHVCFSQVFRERGGLTGIVDYTNYDDVKYAIRKLDDSEFRNAFSRS 581
Query: 181 YIRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSR--SRSRSYSGRSISLSPTAKRSR 238
YIRVREYDR +SRSPSRDSRRSYSR SRSPY+SRSR S S SYSGRS SLSP AK SR
Sbjct: 582 YIRVREYDRSHSRSPSRDSRRSYSR--SRSPYVSRSRSCSLSHSYSGRSRSLSPKAKHSR 755
Query: 239 RLLSLSRCGLLRGSGNCMLGS 259
R SLSRC LLR GNCMLGS
Sbjct: 756 RSFSLSRCSLLRRPGNCMLGS 818
>TC87205 similar to GP|12324445|gb|AAG52185.1 putative splicing factor;
53460-55514 {Arabidopsis thaliana}, partial (81%)
Length = 1197
Score = 294 bits (753), Expect = 2e-80
Identities = 156/244 (63%), Positives = 182/244 (73%), Gaps = 12/244 (4%)
Frame = +2
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAED 60
MSSR SRTIYVGNLP DIR E+ED+FYK+G I++I+LK+PPRPP Y FV+F++ARDAED
Sbjct: 101 MSSRFSRTIYVGNLPADIRESEIEDLFYKYGRIMEIELKVPPRPPCYCFVEFDNARDAED 280
Query: 61 AIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGA------------SRR 108
AI RDGY+FDG RLRVELAHGGRG SSS R GG RG SR
Sbjct: 281 AIRGRDGYNFDGCRLRVELAHGGRGPSSSDRRGYGGGGGGGGRGGGGDSAGGGRFGVSRH 460
Query: 109 SDYRVLVTGLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKL 168
S++RV+V GLP SASWQDLKDHMRKAGDVCF++V R+ G G+VDYTNYDDMKYA+RKL
Sbjct: 461 SEFRVIVRGLPSSASWQDLKDHMRKAGDVCFAEVSRDSEGTFGLVDYTNYDDMKYAIRKL 640
Query: 169 DDSEFRNAFSRAYIRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSGRSI 228
DD+EFRN ++R+YIRVR+Y+ SRS SR RS S +RS L RS SRSRS S RS
Sbjct: 641 DDTEFRNPWARSYIRVRKYESSRSRSRSRSPSRSRSPKRARSRSLERSVSRSRSIS-RSR 817
Query: 229 SLSP 232
S SP
Sbjct: 818 SASP 829
>BF650405 similar to GP|20161426|db putative pre-mRNA splicing factor SF2
{Oryza sativa (japonica cultivar-group)}, partial (49%)
Length = 628
Score = 180 bits (456), Expect(2) = 6e-57
Identities = 88/102 (86%), Positives = 93/102 (90%)
Frame = +3
Query: 27 FYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRDGYDFDGFRLRVELAHGGRGS 86
F+ FGPIVDI+LKIPPRPPGYAFV+FEDARDAEDAI YRDGY FDGFRLRVELAHGGRG
Sbjct: 234 FWVFGPIVDIELKIPPRPPGYAFVEFEDARDAEDAIRYRDGYKFDGFRLRVELAHGGRGY 413
Query: 87 SSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASWQDLK 128
SSSVDRYSSYS G SRG S+ S+YRVLVTGLPPSASWQDLK
Sbjct: 414 SSSVDRYSSYSSG--SRGVSKHSEYRVLVTGLPPSASWQDLK 533
Score = 58.2 bits (139), Expect(2) = 6e-57
Identities = 27/29 (93%), Positives = 28/29 (96%)
Frame = +2
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYK 29
MS RSSRTIYVGNLPGDIRLREVED+FYK
Sbjct: 62 MSRRSSRTIYVGNLPGDIRLREVEDLFYK 148
>TC81507 similar to GP|22831146|dbj|BAC16007. putative pre-mRNA splicing
factor SF2 (SR1 protein) {Oryza sativa (japonica
cultivar-group)}, partial (57%)
Length = 1287
Score = 206 bits (524), Expect = 7e-54
Identities = 101/140 (72%), Positives = 114/140 (81%), Gaps = 1/140 (0%)
Frame = +2
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAED 60
MS SSRT+YVGNLPGDIR REVED+F K+G I IDLK+PPRPPGYAFV+FED +DAED
Sbjct: 92 MSRHSSRTVYVGNLPGDIREREVEDLFMKYGHITHIDLKVPPRPPGYAFVEFEDVQDAED 271
Query: 61 AIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPP 120
AI RDGYDFDG RLRVE AHGGRG+SSS DRYSS+S G RG SRRS+YRV+V GLP
Sbjct: 272 AIRGRDGYDFDGHRLRVEAAHGGRGNSSSRDRYSSHSNGRGGRGVSRRSEYRVIVNGLPS 451
Query: 121 SASWQDLKD-HMRKAGDVCF 139
SASWQDLK+ H + G + F
Sbjct: 452 SASWQDLKESHAKSRGCLLF 511
Score = 144 bits (364), Expect = 3e-35
Identities = 80/115 (69%), Positives = 88/115 (75%), Gaps = 2/115 (1%)
Frame = +3
Query: 127 LKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYIRVRE 186
L+ HMRKAGDVCFSQVF + G TGIVDYTNYDDMKYA++KLDDSEFRNAFS++Y+RVRE
Sbjct: 471 LRSHMRKAGDVCFSQVFHDGRGTTGIVDYTNYDDMKYAIKKLDDSEFRNAFSKSYVRVRE 650
Query: 187 YD-RDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYS-GRSISLSPTAKRSRR 239
YD R SRSP R S RS SRS SRSRS SRSYS G S S SP K S+R
Sbjct: 651 YDSRRDSRSPGRGPSHSRGRSYSRS--RSRSRSHSRSYSPGHSRSKSPKGKSSQR 809
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 85.5 bits (210), Expect = 2e-17
Identities = 85/240 (35%), Positives = 110/240 (45%), Gaps = 9/240 (3%)
Frame = +2
Query: 9 IYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRDGY 68
+YVG L R R++E +F ++G + D+D+K YAFV+F D RDA+DA Y DG
Sbjct: 77 LYVGRLSSRTRSRDLERVFSRYGSVRDVDMK-----HDYAFVEFRDPRDADDARYNLDGR 241
Query: 69 DFDGFRLRVELAHG-GRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASWQDL 127
D DG RL VE A G RGS S D Y G G+ R + + W
Sbjct: 242 DIDGSRLIVEFAKGVPRGSRDSRDS-REYLGRGPPPGSGRCFNCGI-------DGHWA-- 391
Query: 128 KDHMRKAGD---VCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYIRV 184
KAGD C+ R G G ++ + K + +KL S + SR+ R
Sbjct: 392 --RDCKAGDWKNKCY------RCGERGHIE----KNCKNSPKKL--SRHARSVSRSPGRS 529
Query: 185 REYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRSRS-----RSYSGRSISLSPTAKRSRR 239
R R S + SR RR SR S SP S SRSRS RS S SP +SR+
Sbjct: 530 RSPARSRSPARSRSPRRGRSRDRSYSPARSYSRSRSPVRRDRSPVASDRSRSPPPSKSRK 709
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 66.6 bits (161), Expect = 9e-12
Identities = 46/126 (36%), Positives = 60/126 (47%), Gaps = 8/126 (6%)
Frame = +2
Query: 2 SSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDA 61
S+ + +Y+GNL I R++ED F+ FG I + + RPPGYAF+ F+D RDA DA
Sbjct: 26 SAETMTRVYIGNLDSRISERDLEDDFHVFGVIRSV--WVARRPPGYAFIDFDDRRDALDA 199
Query: 62 IYYRDGYDFDGFRLRVELAH--------GGRGSSSSVDRYSSYSGGSSSRGASRRSDYRV 113
I DG + RVEL+H GGRG GG R SD +
Sbjct: 200 IRELDGKN----GWRVELSHNSKTGGGGGGRGGGGG-------GGGGRGRSGGGGSDLKC 346
Query: 114 LVTGLP 119
G P
Sbjct: 347 YXCGEP 364
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 66.6 bits (161), Expect = 9e-12
Identities = 72/235 (30%), Positives = 94/235 (39%), Gaps = 7/235 (2%)
Frame = +3
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAED 60
+++ + +YVGNL + R++ED F FG I + + RPPGYAF+ F+D RDA+D
Sbjct: 27 ITNETMSRVYVGNLDSRVSERDLEDEFRVFGVIRSV--WVARRPPGYAFIDFDDRRDAQD 200
Query: 61 AIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSR----GASRRSDYRVLVT 116
AI DG + RVEL+H R GG R G SD +
Sbjct: 201 AIRDLDGKN----GWRVELSHNSRSGGGG-------GGGGGGRGRGGGGGGGSDLKCYEC 347
Query: 117 GLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNA 176
G P F++ R RGG A R+ S R
Sbjct: 348 GEPGH-----------------FARECRNRGG-------------GGAGRRRSRSPPRFR 437
Query: 177 FSRAYIRVREYDRDYS---RSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSGRSI 228
S +Y R R YS RSP R S RS SRSP R R +G +
Sbjct: 438 RSPSYGR-----RSYSPRGRSPRRRSVSPRGRSYSRSPPPYRGREEVPYANGNGV 587
>TC78771 similar to GP|16612268|gb|AAL27502.1 AT3g61860/F21F14_30
{Arabidopsis thaliana}, partial (87%)
Length = 1254
Score = 64.7 bits (156), Expect = 3e-11
Identities = 74/259 (28%), Positives = 104/259 (39%), Gaps = 45/259 (17%)
Frame = +1
Query: 7 RTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRD 66
R ++ GNL D R E+E +F K+G I +D+K G+AFV +ED RDAE+AI D
Sbjct: 91 RPVFAGNLEYDTRQSELERLFSKYGRIERVDMK-----SGFAFVYYEDERDAEEAIRALD 255
Query: 67 GYDF--DGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASW 124
F D RL VE A G RG S+ + + ++ P
Sbjct: 256 NIPFGHDKRRLSVEWARGERGRHR-----------DGSKPNQKPTKTLFVINFDPIRTRV 402
Query: 125 QDLKDHMRKAGDVCFSQV--------FRERGGMTGIVDYTNYD-------DMKYALRKLD 169
D++ H + G + ++ + + T ++ TN ++YALR D
Sbjct: 403 SDIERHFKPYGPLHHVRIRRNFAFVQYETQEDATKALECTNMSKILDRVVSVEYALR--D 576
Query: 170 DSE----------------------FRNAFSRAYIRVRE--YDR----DYSRSPSRDSRR 201
DS+ +R S Y R R YDR D RSP R
Sbjct: 577 DSDRVDNYGGSPRRGGGLARSPSPGYRRRPSPDYGRPRSPVYDRYTGPDRRRSPDYGRNR 756
Query: 202 SYSRSISRSPYLSRSRSRS 220
S +RSP R RSRS
Sbjct: 757 SPDYGRNRSPEYGRYRSRS 813
>BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (54%)
Length = 364
Score = 62.0 bits (149), Expect = 2e-10
Identities = 38/99 (38%), Positives = 54/99 (54%)
Frame = +1
Query: 9 IYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRDGY 68
+Y+GNL + RE+ED F +G + + + RPPGYAF++F+D RDA DAI+ DG
Sbjct: 49 VYIGNLDPRVTERELEDEFRVYGVLRSV--WVARRPPGYAFIEFDDPRDALDAIHELDGK 222
Query: 69 DFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASR 107
+ RV+L+H +S SGG RG R
Sbjct: 223 N----GWRVQLSH------------NSKSGGGGGRGGGR 291
>TC76468 similar to PIR|E84638|E84638 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (39%)
Length = 465
Score = 61.6 bits (148), Expect = 3e-10
Identities = 35/83 (42%), Positives = 49/83 (58%)
Frame = +3
Query: 2 SSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDA 61
S+ + +Y+GNL I R++ED F+ FG I + + RPPGYAF+ F+D RDA DA
Sbjct: 156 SAETMTRVYIGNLDSRISERDLEDDFHVFGVIRSV--WVARRPPGYAFIDFDDRRDALDA 329
Query: 62 IYYRDGYDFDGFRLRVELAHGGR 84
I DG + RVEL+H +
Sbjct: 330 IRELDGKN----GWRVELSHNSK 386
>TC83750 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (68%)
Length = 772
Score = 60.8 bits (146), Expect = 5e-10
Identities = 44/114 (38%), Positives = 58/114 (50%), Gaps = 3/114 (2%)
Frame = +2
Query: 2 SSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDA 61
S+ + +Y+GNL I R++ED F+ FG I + + RPPGYAF+ F+D RDA DA
Sbjct: 32 SAETMTRVYIGNLDSRISERDLEDDFHVFGVIRSV--WVARRPPGYAFIDFDDRRDALDA 205
Query: 62 IYYRDGYDFDGFRLRVELAHGGRG--SSSSVDRY-SSYSGGSSSRGASRRSDYR 112
I DG+ F + GG G S S R+ S S G S RS R
Sbjct: 206 IRELDGH----FARECNSSRGGSGRRRSRSPPRFRRSPSYGRRSYSPRERSPRR 355
Score = 29.3 bits (64), Expect = 1.6
Identities = 44/152 (28%), Positives = 56/152 (35%), Gaps = 25/152 (16%)
Frame = +2
Query: 112 RVLVTGLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDS 171
RV + L S +DL+D G + V R G +D+ + D A+R+LD
Sbjct: 50 RVYIGNLDSRISERDLEDDFHVFGVIRSVWVARRPPGYA-FIDFDDRRDALDAIRELDGH 226
Query: 172 EFRNAFSRAYIRVREYDRD---YSRSPSRDSRRSYS----------------------RS 206
R S R R + RSPS RRSYS RS
Sbjct: 227 FARECNSSRGGSGRRRSRSPPRFRRSPSY-GRRSYSPRERSPRRRSPSPRRRSPSPRRRS 403
Query: 207 ISRSPYLSRSRSRSRSYSGRSISLSPTAKRSR 238
SRSP R R + +G I A RSR
Sbjct: 404 YSRSPPPYRGREDATHANGNGIR---AAHRSR 490
>AL368496 similar to GP|16612268|gb| AT3g61860/F21F14_30 {Arabidopsis
thaliana}, partial (36%)
Length = 616
Score = 58.2 bits (139), Expect = 3e-09
Identities = 36/78 (46%), Positives = 45/78 (57%), Gaps = 2/78 (2%)
Frame = -2
Query: 10 YVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRDGYD 69
+ GNL D R E+E +F K+G I +D+K G+AFV +ED RDAE+AI D
Sbjct: 594 FAGNLEYDTRQSELERLFSKYGRIERVDMK-----SGFAFVYYEDERDAEEAIRALDNIP 430
Query: 70 F--DGFRLRVELAHGGRG 85
F D RL VE A G RG
Sbjct: 429 FGHDKRRLSVEWARGERG 376
>TC84230
Length = 691
Score = 54.3 bits (129), Expect = 5e-08
Identities = 45/165 (27%), Positives = 72/165 (43%)
Frame = +2
Query: 2 SSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDA 61
SS S +YV NL D+ ++ D+F ++G + D YAFV F+ DA+ A
Sbjct: 203 SSTPSNNLYVANLSPDVTDSDLMDLFVQYGAL---DSVTSYSARNYAFVFFKRIDDAKAA 373
Query: 62 IYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPS 121
G++F G LR+E A + ++ V G+ P+
Sbjct: 374 KNALQGFNFRGNSLRIEFARPAKTCK------------------------QLWVGGISPA 481
Query: 122 ASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALR 166
+ +DL+ RK G V + FR+R T V++ N DD A++
Sbjct: 482 VTKEDLEADFRKFGKVEDFKFFRDRN--TACVEFFNLDDAIQAMK 610
>TC87334 similar to GP|15293081|gb|AAK93651.1 unknown protein {Arabidopsis
thaliana}, partial (54%)
Length = 977
Score = 52.8 bits (125), Expect = 1e-07
Identities = 44/108 (40%), Positives = 53/108 (48%), Gaps = 8/108 (7%)
Frame = +2
Query: 144 RERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRA-----YIRVREYDRDYSRSPSRD 198
RERG +YD + R+ + +SR+ R R Y R S R
Sbjct: 515 RERG--------RSYDYRRSPRRRSRSPRYARTYSRSPDYTPSPRPRRYSRSISPRDERY 670
Query: 199 SRRSYSRSISRSPYLSRSRSRSRSYSGRSISLSPTAKR---SRRLLSL 243
RRSYSRS RSPY SRS R RSYS RSIS SP R S RL+++
Sbjct: 671 RRRSYSRSPYRSPYGSRSPDRGRSYS-RSISRSPGYSR*FKSTRLVNM 811
>TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (18%)
Length = 1681
Score = 51.2 bits (121), Expect = 4e-07
Identities = 56/237 (23%), Positives = 96/237 (39%), Gaps = 36/237 (15%)
Frame = -2
Query: 2 SSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPR--PPGYAFVQFEDARDAE 59
+S+ R +YVGNL D++ ++D + G ++ D+ + P G V++ A+
Sbjct: 1374 TSQQDRRVYVGNLSYDVKWHHLKDFMRQAGEVLFADVLLLPNGMSKGCGIVEYATREQAQ 1195
Query: 60 DAI-------------YYRDGYDFDGFRLRVELAHGGRGSSSS------------VDRYS 94
+A+ Y R+ + + + GG G +
Sbjct: 1194 NAVATLSNQNLMGRLVYVREDREAEPRFIGATANRGGFGGGPGGPGGPGGPGGMHPGGFG 1015
Query: 95 SYSGGSSSRGASRRSDYRVLVTGLPPSASWQDLKDHMRKA--------GDVCFSQVFRER 146
GS S G SR+ + V LP + WQDLKD R+A DV R +
Sbjct: 1014 GGGPGSYSTGGSRQ----IFVANLPYTVGWQDLKDLFRQAARNGVVIRADVHLGPDGRPK 847
Query: 147 GGMTGIVDYTNYDDMKYALRKLDDSEFR-NAFSRAYIRVREYDRDYSRSPSRDSRRS 202
G +GIV + N +D + A+++ +++ A A ++ R Y SR RR+
Sbjct: 846 G--SGIVVFENPEDARNAIQQFHGYDWQGRAAGSARRQICRRRRPYGFWWSRRLRRN 682
Score = 42.7 bits (99), Expect = 1e-04
Identities = 30/97 (30%), Positives = 43/97 (43%), Gaps = 2/97 (2%)
Frame = -1
Query: 8 TIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPP--RPPGYAFVQFEDARDAEDAIYYR 65
TIYV NLP ++ ++F G + +++ P R G V+F+ A AE AI
Sbjct: 484 TIYVRNLPWSTSNEDLVELFTTIGKVEQAEIQYEPSGRSRGTGVVRFDSAETAETAIAKF 305
Query: 66 DGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSS 102
GY + GGR S RY + +GG S
Sbjct: 304 QGYQY-----------GGRPLGLSFVRYLNAAGGGDS 227
Score = 31.2 bits (69), Expect = 0.42
Identities = 19/39 (48%), Positives = 22/39 (55%), Gaps = 1/39 (2%)
Frame = -2
Query: 189 RDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSR-SYSGR 226
R+ SRSP R RS + R + RSRSRSR Y GR
Sbjct: 1626 RERSRSPRRGRSRSPTNGPRRRSFSPRSRSRSRDDYRGR 1510
Score = 27.3 bits (59), Expect = 6.0
Identities = 23/48 (47%), Positives = 25/48 (51%), Gaps = 6/48 (12%)
Frame = -2
Query: 198 DSRRSYS-RSISRSPYLSRSRS-----RSRSYSGRSISLSPTAKRSRR 239
DS +Y R SRSP RSRS R RS+S RS S S R RR
Sbjct: 1650 DSGDNYRPRERSRSPRRGRSRSPTNGPRRRSFSPRSRSRSRDDYRGRR 1507
>TC84024 similar to PIR|S77714|S77714 RNA-binding protein precursor 33K -
wood tobacco, partial (55%)
Length = 867
Score = 50.8 bits (120), Expect = 5e-07
Identities = 44/181 (24%), Positives = 81/181 (44%), Gaps = 10/181 (5%)
Frame = +3
Query: 2 SSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDL---KIPPRPPGYAFVQFEDARDA 58
+S + +YVGNLP + ++ DIF + G +V +++ ++ R G+AFV + A +A
Sbjct: 318 TSDDAGRLYVGNLPFSMTSSQLADIFAEAGTVVSVEIVYDRLTDRSRGFAFVTMKSAEEA 497
Query: 59 EDAIYYRDGYDFDGFRLRV---ELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLV 115
++AI DG G +V E+ GG +SY G S +++
Sbjct: 498 KEAIRMFDGSQVGGRSAKVNFPEVPKGGERLVMGQKVRNSYRGFVD-------SPHKIYA 656
Query: 116 TGLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMT----GIVDYTNYDDMKYALRKLDDS 171
L + QDL+D + + +++ ER G V + + ++ AL ++
Sbjct: 657 GNLGWGMNSQDLRDAFDEQPGLLGAKIIYERDNGRSRGFGFVTFETAEHLEAALNAMNXV 836
Query: 172 E 172
E
Sbjct: 837 E 839
>TC76948 similar to SP|Q08935|ROC1_NICSY 29 kDa ribonucleoprotein A
chloroplast precursor (CP29A). [Wood tobacco] {Nicotiana
sylvestris}, partial (61%)
Length = 1374
Score = 50.4 bits (119), Expect = 7e-07
Identities = 47/179 (26%), Positives = 78/179 (43%), Gaps = 12/179 (6%)
Frame = +1
Query: 6 SRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDL---KIPPRPPGYAFVQFEDARDAEDAI 62
++ ++VGNLP + ++ +IF G + +++ K R G+ FV A + E A
Sbjct: 337 NQRLFVGNLPFSVDSAQLAEIFENAGDVEMVEVIYDKSTGRSRGFGFVTMSSAAEVEAAA 516
Query: 63 YYRDGYDFDGFRLRVELA--HGGRGSSSSVDRYSSYSGG---SSSRGASRRSDYRVLVTG 117
+GY DG LRV R +S + G RG S D RV V
Sbjct: 517 QQLNGYVVDGRELRVNAGPPPPPRSENSRFGENPRFGGDRPRGPPRGGSSDGDNRVHVGN 696
Query: 118 LPPSASWQDLKDHMRKAGDVCFSQVFRER-GGMT---GIVDYTNYDDMKYALRKLDDSE 172
L L+ + G V ++V +R G + G V +++ D++ A+R LD ++
Sbjct: 697 LAWGVDNLALESLFGEQGQVLEAKVIYDRESGRSRGFGFVTFSSADEVDSAIRTLDGAD 873
>TC83951 similar to GP|22136316|gb|AAM91236.1 putative RNA binding protein
{Arabidopsis thaliana}, partial (47%)
Length = 659
Score = 49.3 bits (116), Expect = 1e-06
Identities = 59/212 (27%), Positives = 82/212 (37%), Gaps = 5/212 (2%)
Frame = +3
Query: 18 IRLREVEDIFYKFGPIVDIDLK---IPPRPPGYAFVQFEDARDAEDAIYYRDGYDFDGFR 74
+ LR E F +FGP+ D+ + +P G+AFVQF DA +A +A Y+ D F G
Sbjct: 48 VDLRS*EPPFERFGPVRDVYIPKDYYSGQPRGFAFVQFVDAYEASEAQYHMDRQIFAGRE 227
Query: 75 LRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASWQDLKDHMRKA 134
+ V +A R + +S S G +RS Y H R
Sbjct: 228 ISVVVAAETRKRPEEMRHRTSRSRGPGGSYGGQRSSY------------------HGRSR 353
Query: 135 GDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYIRVREYDRDYSRS 194
+ R R Y + +Y R SR++ DYS S
Sbjct: 354 S----RSISRSRS-----PPYHSGSRNRYRSR-----------SRSFSPAPRRQGDYSVS 473
Query: 195 PSRDSRRSYS-RSISRSPYLSRSRS-RSRSYS 224
P R + R S RS RSP + R + RSYS
Sbjct: 474 PRRHAERPRSPRSPPRSPPVERDADHKRRSYS 569
Score = 32.0 bits (71), Expect = 0.24
Identities = 23/39 (58%), Positives = 25/39 (63%), Gaps = 3/39 (7%)
Frame = +3
Query: 201 RSYSRSISRS---PYLSRSRSRSRSYSGRSISLSPTAKR 236
RS SRSISRS PY S SR+R RS RS S SP +R
Sbjct: 345 RSRSRSISRSRSPPYHSGSRNRYRS---RSRSFSPAPRR 452
>TC79595 similar to GP|3850823|emb|CAA77136.1 U2 snRNP auxiliary factor
large subunit {Nicotiana plumbaginifolia}, partial (77%)
Length = 1525
Score = 49.3 bits (116), Expect = 1e-06
Identities = 35/81 (43%), Positives = 43/81 (52%)
Frame = +2
Query: 178 SRAYIRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSGRSISLSPTAKRS 237
SR Y R REYDR+ R SR RS SRS +RS + SRSRSRSRS S R+ S
Sbjct: 8 SRGYDRRREYDRE-DRHRSRRRSRSRSRSRARSEHRSRSRSRSRSKSKRTSGFDMAPPTS 184
Query: 238 RRLLSLSRCGLLRGSGNCMLG 258
L + G + G+ + G
Sbjct: 185 AILGATGVAGQITGASPAIPG 247
Score = 26.9 bits (58), Expect = 7.8
Identities = 14/49 (28%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Frame = +2
Query: 9 IYVGNLPGDIRLREVEDIFYKFGPIVDIDL---KIPPRPPGYAFVQFED 54
I+VG +P ++ ++ FGP+ DL + GYAF ++D
Sbjct: 719 IFVGGVPYYFTETQIRELLETFGPLRGFDLVKDRETGNSKGYAFCVYQD 865
>TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-rich
splicing factor - alfalfa, partial (95%)
Length = 1494
Score = 48.1 bits (113), Expect = 3e-06
Identities = 65/238 (27%), Positives = 96/238 (40%), Gaps = 25/238 (10%)
Frame = +3
Query: 26 IFYKFGPIVDIDLKIPPRPP---GYAFVQFEDARDAEDAIYYRDGYDFDGFRLRVELAHG 82
+F K+G +VDI + R G+AFV+++ A +A A+ DG DG + V+ A
Sbjct: 141 LFDKYGKVVDIFIPKDRRTGESRGFAFVRYKYADEASKAVDRLDGRMVDGREITVQFAKY 320
Query: 83 G----RGSSSSVDRYSSYSGGSSSRGASRRS---------DYRVLVTGLPPSASWQDL-- 127
G R + S S S SR S+RS DYR +D
Sbjct: 321 GPNAERIQKGRIIETSPRSKSSRSRSPSKRSRHRDDYKEKDYRRRSRSRSYDRHERDRHR 500
Query: 128 ---KDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYIRV 184
+DH R++ S ++ RG +DD + + + R+ R+ I
Sbjct: 501 GRDRDHRRRSRSRSASPGYKGRGR-------GRHDDERRSRSPSRSVDSRSPVRRSSIPK 659
Query: 185 REYDRDYSRSPSRDS--RRSYSRSISRSPYLS--RSRSRSRSYSGRSISLSPTAKRSR 238
R S SP R +RS S S SP S R +RS GRS++ + R R
Sbjct: 660 RSPSPKRSPSPKRSPSLKRSISPQKSVSPRKSPLRESPDNRSRGGRSLTPRSVSPRGR 833
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,237,503
Number of Sequences: 36976
Number of extensions: 95670
Number of successful extensions: 1140
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 764
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 918
length of query: 259
length of database: 9,014,727
effective HSP length: 94
effective length of query: 165
effective length of database: 5,538,983
effective search space: 913932195
effective search space used: 913932195
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0299.13


