

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0290.23
(358 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
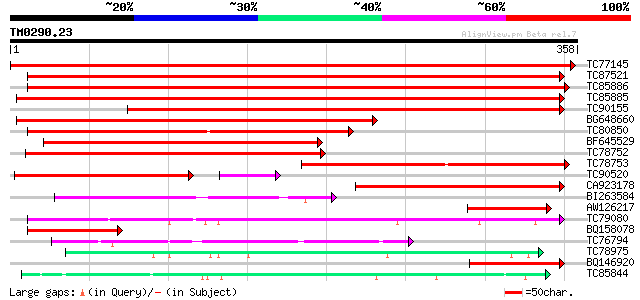
Score E
Sequences producing significant alignments: (bits) Value
TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehyd... 648 0.0
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 367 e-102
TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 365 e-101
TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 362 e-101
TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 272 2e-73
BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogena... 243 1e-64
TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehyd... 227 6e-60
BF645529 weakly similar to GP|22475166|gb putative sinapyl alcoh... 184 3e-47
TC78752 similar to GP|22857590|gb|AAN09864.1 putative cinnamyl-a... 184 3e-47
TC78753 similar to PIR|E96751|E96751 hypothetical protein F28P22... 147 5e-36
TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol deh... 129 5e-33
CA923178 weakly similar to SP|P93257|MTD_ Probable mannitol dehy... 126 1e-29
BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1... 109 2e-24
AW126217 homologue to GP|17028158|gb| cinnamyl-alcohol dehydroge... 100 7e-22
TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogena... 99 3e-21
BQ158078 similar to GP|10187159|emb cDNA~Strawberry alcohol dehy... 80 2e-15
TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbi... 74 9e-14
TC78975 similar to GP|21592656|gb|AAM64605.1 alcohol dehydrogena... 65 5e-11
BQ146920 similar to GP|8099340|gb|A ELI3 {Lycopersicon esculentu... 64 1e-10
TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 62 3e-10
>TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) (CAD). [Alfalfa] {Medicago sativa},
complete
Length = 1384
Score = 648 bits (1671), Expect = 0.0
Identities = 313/358 (87%), Positives = 337/358 (93%), Gaps = 1/358 (0%)
Frame = +1
Query: 1 MGSLE-AERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLG 59
MGS+E AERTTVG AA+DPSGIL+PYT+TLRNTGPDDVYIK+HYCGVCH+D+HQ+KNDLG
Sbjct: 52 MGSIEVAERTTVGLAAKDPSGILTPYTYTLRNTGPDDVYIKIHYCGVCHSDLHQIKNDLG 231
Query: 60 MSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIW 119
MSNYPMVPGHEVVGEVLEVGS+VTRF VGEIVG GLLVGCCK C AC S+IEQYC KKIW
Sbjct: 232 MSNYPMVPGHEVVGEVLEVGSNVTRFKVGEIVGVGLLVGCCKSCRACDSEIEQYCNKKIW 411
Query: 120 NYNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKE 179
+YNDVY DGK TQGGFAE+ +VEQKFVVKIPEG+APEQVAPLLCA VTVYSPLSHFGLK
Sbjct: 412 SYNDVYTDGKITQGGFAESTVVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLSHFGLKT 591
Query: 180 SGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQ 239
GLRGGILGLGGVGHMGV +AKA GHHVTVISSSD+KKKEA+EDLGAD+YLVSSDT MQ
Sbjct: 592 PGLRGGILGLGGVGHMGVKVAKAFGHHVTVISSSDKKKKEALEDLGADSYLVSSDTVGMQ 771
Query: 240 EAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSF 299
EAADSLDYIIDTVPVGHPLEPYLSLLK+DGKLILMGVINTPLQF+TPMVMLGR+SITGSF
Sbjct: 772 EAADSLDYIIDTVPVGHPLEPYLSLLKIDGKLILMGVINTPLQFVTPMVMLGRKSITGSF 951
Query: 300 IGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGSKLID 357
+GS+KETEEMLEFWKEKGL+SMIEIV MDYINKAFERLEKNDVRYRFVVDVKGSK D
Sbjct: 952 VGSVKETEEMLEFWKEKGLSSMIEIVTMDYINKAFERLEKNDVRYRFVVDVKGSKFED 1125
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 367 bits (943), Expect = e-102
Identities = 172/339 (50%), Positives = 243/339 (70%)
Frame = +2
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GWAARD SG+LSP+ F+ R TG DV KV YCG+CH+D+H +KN+ GMS YP+VPGHE+
Sbjct: 110 GWAARDTSGVLSPFNFSRRETGEKDVAFKVLYCGICHSDLHMIKNEWGMSTYPLVPGHEI 289
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPT 131
G V EVGS V +F +G+ VG G LV C+ C C+ ++E YC K+ Y+ Y DG T
Sbjct: 290 AGIVTEVGSKVEKFKIGDKVGVGCLVDSCRACQNCEENLENYCPKQTNTYSAKYSDGSIT 469
Query: 132 QGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGG 191
GG++++++ ++ F+V IP+G+ E APLLCA +TVYSPL +FGL + G+ GI+GLGG
Sbjct: 470 YGGYSDSMVADEHFIVHIPDGLPLESAAPLLCAGITVYSPLRYFGLDKPGMNIGIVGLGG 649
Query: 192 VGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDT 251
+GH+GV AKA G +VTVIS+S K+KEAIE+LGAD++L+S D MQ A +LD IIDT
Sbjct: 650 LGHLGVKFAKAFGANVTVISTSPNKEKEAIENLGADSFLISHDQDKMQAAMGTLDGIIDT 829
Query: 252 VPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLE 311
V HPL P + LLK GKL+++G + P + +++GR++I+GS IG +KET+EM++
Sbjct: 830 VSADHPLLPLVGLLKYHGKLVMVGAPDKPPELPHIPLIMGRKTISGSGIGGMKETQEMID 1009
Query: 312 FWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
F + + IE++ +DY+N A +RL K DV+YRFV+D+
Sbjct: 1010FAAKHNIKPDIEVIPVDYVNTAMKRLLKADVKYRFVLDI 1126
>TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (94%)
Length = 1395
Score = 365 bits (936), Expect = e-101
Identities = 175/342 (51%), Positives = 237/342 (69%)
Frame = +2
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GWAARD SG+LSP+ F R TG DV KV YCG+CH+D+H ++N+ G + YP+VPGHE+
Sbjct: 113 GWAARDSSGVLSPFHFFRRETGEKDVAFKVLYCGICHSDLHIMQNEWGKTTYPLVPGHEL 292
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPT 131
G V EVGS V +F VG+ VG G +V C+ C C DIE YC K +N DG T
Sbjct: 293 TGVVTEVGSKVKKFKVGDKVGVGYMVDSCRSCENCADDIENYCTKYTQTFNGKSRDGTIT 472
Query: 132 QGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGG 191
GGF+++++ ++ FV++IP+ + + PLLCA VTVYSPL HFGL + G+ G++GLGG
Sbjct: 473 YGGFSDSMVADEHFVIRIPDSLPLDGAGPLLCAGVTVYSPLRHFGLDKPGMNIGVVGLGG 652
Query: 192 VGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDT 251
+GHM V AKA G VTVIS+S +K+KEAIE LGAD++LVS D MQ A +L+ IIDT
Sbjct: 653 LGHMAVKFAKAFGAKVTVISTSPKKEKEAIEHLGADSFLVSRDPEQMQAATSTLNGIIDT 832
Query: 252 VPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLE 311
V HP+ P + LLK +GKL+++G + PL+ ++ GR+SI GS IG IKET+EM++
Sbjct: 833 VSASHPVVPLIGLLKSNGKLVMVGAVAKPLELPIFSLLGGRKSIAGSLIGGIKETQEMID 1012
Query: 312 FWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGS 353
F + +T IE+V +DY+N A ERL K DV+YRFV+D+ S
Sbjct: 1013FAAKHNVTPEIEVVPIDYVNTAMERLVKGDVKYRFVIDIGNS 1138
>TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (97%)
Length = 1330
Score = 362 bits (930), Expect = e-101
Identities = 174/346 (50%), Positives = 238/346 (68%)
Frame = +3
Query: 5 EAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYP 64
E + GWAARD SG+LSP+ F+ R TG DV KV YCG+CHTD+H +KN+ G S YP
Sbjct: 90 EHPKKAFGWAARDSSGVLSPFNFSRRETGEKDVAFKVLYCGICHTDLHMMKNEWGNSIYP 269
Query: 65 MVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDV 124
+VPGHE+ G V EVGS V +F VG+ VG G +V C+ C C D+E YC ++
Sbjct: 270 LVPGHELAGIVTEVGSKVEKFKVGDKVGVGYMVDSCRSCENCAEDLENYCPQQTVTCGAK 449
Query: 125 YVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRG 184
Y DG T GG++++++ ++ FV++IP+ + + PLLCA VTVYSPL HF L + G+
Sbjct: 450 YRDGSVTYGGYSDSMVADEHFVIRIPDSLPLDVAGPLLCAGVTVYSPLRHFQLDKPGMNI 629
Query: 185 GILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADS 244
G++GLGG+GHM V AKA G +VTVIS+S K+KEAIE LGAD++LVS D MQ A +
Sbjct: 630 GVVGLGGLGHMAVKFAKAFGANVTVISTSPSKEKEAIEHLGADSFLVSRDPDQMQAAMGT 809
Query: 245 LDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIK 304
L+ IIDTV HP+ P + LLK +GKL+++G + PL+ ++ GR+ + GS IG IK
Sbjct: 810 LNGIIDTVSASHPILPLIGLLKSNGKLVMVGGVAKPLELPVFSLLGGRKLVAGSLIGGIK 989
Query: 305 ETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
ET+EM++F E +T IE+V +DY+N A ERLEK DV+YRFV+D+
Sbjct: 990 ETQEMIDFAAEHNVTPDIEVVPIDYVNTAMERLEKADVKYRFVIDI 1127
>TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (78%)
Length = 1051
Score = 272 bits (695), Expect = 2e-73
Identities = 136/276 (49%), Positives = 186/276 (67%)
Frame = +2
Query: 75 VLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGG 134
V EVGS V +F VG+ VG G L+ C+ C C ++E YC K + Y DG T GG
Sbjct: 11 VTEVGSKVEKFKVGDKVGVGYLIDSCRSCQDCNDNLENYCPKFVVTCGAKYRDGTVTYGG 190
Query: 135 FAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGH 194
++++++ ++ FV++IP+ + E PLLCA VTVYSPL FGL + GL G++GLGG+GH
Sbjct: 191 YSDSMVADEHFVIRIPDNIPLEFAGPLLCAGVTVYSPLRFFGLDKPGLHIGVVGLGGLGH 370
Query: 195 MGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPV 254
M V AKA G +VTVIS+S K+KEAIE LGAD++L+SSD +Q A +LD IIDTV
Sbjct: 371 MAVKFAKAFGANVTVISTSPNKEKEAIEHLGADSFLISSDPKQIQGAIGTLDGIIDTVSA 550
Query: 255 GHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLEFWK 314
HPL P + LLK GKLI++GVI PLQ ++ GR+ + GS +G +KET+EM+ F
Sbjct: 551 VHPLLPMIGLLKSHGKLIMLGVIVQPLQLPEYTLIQGRKILAGSQVGGLKETQEMINFAA 730
Query: 315 EKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
E + IE+V +DY+N A +RL K DV+YRFV+D+
Sbjct: 731 EHNVKPDIEVVPIDYVNTAMQRLAKGDVKYRFVIDI 838
>BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (64%)
Length = 721
Score = 243 bits (619), Expect = 1e-64
Identities = 112/228 (49%), Positives = 156/228 (68%)
Frame = +1
Query: 5 EAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYP 64
E + GWAARD SG+LSP+ F+ R DV ++V YCG+CH+D+H+ KN+ G +NYP
Sbjct: 37 EHSKKAFGWAARDSSGVLSPFNFSRREICEKDVALRVLYCGICHSDLHKAKNEWGTTNYP 216
Query: 65 MVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDV 124
+VPGHE+VG V EVGS V +F VG+ VG G LV C C C ++E YC +
Sbjct: 217 LVPGHEIVGIVTEVGSKVKKFKVGDRVGVGCLVDSCHSCQNCVDNLENYCPQLTLTDGSK 396
Query: 125 YVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRG 184
Y DG T GGF+++++ ++ FV +IP+ + + APLLCA +TVYSPL HFGL + +
Sbjct: 397 YSDGTSTHGGFSDSMVSDEHFVFRIPDQLPLDAAAPLLCAGITVYSPLRHFGLDKPDMNI 576
Query: 185 GILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVS 232
G++GLGG+GHM V AKA G +VTVIS+S +K+ EA+E L AD++L+S
Sbjct: 577 GVVGLGGLGHMAVKFAKAFGANVTVISTSPKKENEALEHLRADSFLIS 720
>TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Alfalfa], partial (61%)
Length = 710
Score = 227 bits (578), Expect = 6e-60
Identities = 101/206 (49%), Positives = 144/206 (69%)
Frame = +1
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GWAARD SG LSP+ F+ R G DDV +K+ YCGVCH+D+H +KND G + YP+VPGHE+
Sbjct: 94 GWAARDTSGTLSPFHFSRRENGDDDVSVKILYCGVCHSDLHTLKNDWGFTTYPVVPGHEI 273
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPT 131
VG V +VG +V +F VG+ VG G++V C+ C C D+EQYC K ++ YN Y G T
Sbjct: 274 VGVVTKVGINVKKFRVGDNVGVGVIVESCQTCENCNQDLEQYCPKLVFTYNSPY-KGTRT 450
Query: 132 QGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGG 191
GG+++ ++V Q++VV+ P+ + + APLLCA +TVYSP+ ++G+ E G G+ GLGG
Sbjct: 451 HGGYSDFVVVHQRYVVQFPDNLPLDAGAPLLCAGITVYSPMKYYGMTEPGKHLGVAGLGG 630
Query: 192 VGHMGVIIAKAMGHHVTVISSSDRKK 217
+GH+ + KA G VTVI++S K+
Sbjct: 631 LGHVAIKFGKAFGLKVTVITTSPNKE 708
>BF645529 weakly similar to GP|22475166|gb putative sinapyl alcohol
dehydrogenase {Populus tremula x Populus tremuloides},
partial (40%)
Length = 658
Score = 184 bits (468), Expect = 3e-47
Identities = 84/177 (47%), Positives = 119/177 (66%), Gaps = 1/177 (0%)
Frame = +3
Query: 22 LSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEVVGEVLEVGSD 81
++PYTF R G DV IK+ YCG+CHTDVH K+D G++ YP+VPGHE+ G + +VGSD
Sbjct: 99 VTPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSD 278
Query: 82 VTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIV 141
V F G+ VG G L C C C++D E YC+K + YN ++ DG T GG+++ ++V
Sbjct: 279 VEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSITYGGYSQMLVV 458
Query: 142 EQKFVVKIPEGMAPEQVAPLLCAAVTVYSPL-SHFGLKESGLRGGILGLGGVGHMGV 197
+ ++VV IPE + + APLLCA +TV+SPL H + +G R G++GLG +GHM V
Sbjct: 459 DYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHXLVSTAGKRIGVVGLGXLGHMAV 629
>TC78752 similar to GP|22857590|gb|AAN09864.1 putative cinnamyl-alcohol
dehydrogenase {Oryza sativa (japonica cultivar-group)},
partial (50%)
Length = 668
Score = 184 bits (468), Expect = 3e-47
Identities = 85/190 (44%), Positives = 120/190 (62%), Gaps = 1/190 (0%)
Frame = +2
Query: 11 VGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHE 70
+ WAARD SG+LSPY F R G +DVY+K+ +CGVC+ DV KN G S YP+VPGHE
Sbjct: 74 LAWAARDASGVLSPYKFNRRELGSEDVYVKITHCGVCYADVIWAKNKHGDSKYPVVPGHE 253
Query: 71 VVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYC-KKKIWNYNDVYVDGK 129
+ G V +VG +V RF VG+ VG G + C++C C E +C K ++ +N V DG
Sbjct: 254 IAGVVAKVGPNVQRFKVGDHVGVGTYINSCRECEYCNDRFEVHCVKGSVYTFNGVDYDGT 433
Query: 130 PTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGL 189
T+GG++ +I+V +++ IP+ PLLCA +TVYSP+ + + G G++GL
Sbjct: 434 ITKGGYSTSIVVHERYCFLIPKSHPLASAGPLLCAGITVYSPMIRHNMNQPGKSLGVVGL 613
Query: 190 GGVGHMGVII 199
GG+GHM V I
Sbjct: 614 GGLGHMAVKI 643
>TC78753 similar to PIR|E96751|E96751 hypothetical protein F28P22.13
[imported] - Arabidopsis thaliana, partial (47%)
Length = 865
Score = 147 bits (372), Expect = 5e-36
Identities = 76/169 (44%), Positives = 111/169 (64%)
Frame = +3
Query: 185 GILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADS 244
G++GLGG+GHM V KA G VTV S+S KK+EA+ LGAD ++VSS+ M+ A S
Sbjct: 3 GVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSLLGADQFVVSSNQEDMRALAKS 182
Query: 245 LDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIK 304
LD+IIDT H +PY+SLLK+ G L+L+G + ++F + LG R++ GS G K
Sbjct: 183 LDFIIDTASGDHLFDPYMSLLKISGVLVLVG-FPSEVKFSPASLNLGSRTVAGSVTGGTK 359
Query: 305 ETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGS 353
E +EM++F G+ IE++ + Y N+A ER+ DV+YRFV+D++ S
Sbjct: 360 EIQEMVDFCAANGIHPDIELIPIGYSNEALERVVNKDVKYRFVIDIENS 506
>TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) CAD1 - Arabidopsis thaliana, partial
(41%)
Length = 618
Score = 129 bits (325), Expect(2) = 5e-33
Identities = 57/113 (50%), Positives = 73/113 (64%)
Frame = +2
Query: 4 LEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNY 63
L + GWAA D SG ++PYTF R G DV IK+ YCG+CHTDVH K+D G++ Y
Sbjct: 53 LTTHKXVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMY 232
Query: 64 PMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKK 116
P+VPGHE+ G + +VGSDV F G+ VG G L C C C++D E YC+K
Sbjct: 233 PVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEK 391
Score = 29.3 bits (64), Expect(2) = 5e-33
Identities = 14/39 (35%), Positives = 20/39 (50%)
Frame = +1
Query: 133 GGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSP 171
G A ++ K I E + + APLLCA +TV+ P
Sbjct: 421 GMVASPMVATHKCWS*ITESLPMDAAAPLLCAGITVFXP 537
>CA923178 weakly similar to SP|P93257|MTD_ Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).,
partial (34%)
Length = 668
Score = 126 bits (316), Expect = 1e-29
Identities = 64/132 (48%), Positives = 90/132 (67%)
Frame = -3
Query: 219 EAIEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVIN 278
EA E LGAD +++S++ +Q A SLD+I+DTV H L P L LLK++G L ++G +
Sbjct: 663 EAKERLGADDFIISTNPDQLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPD 484
Query: 279 TPLQFITPMVMLGRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLE 338
PLQ ++ G+RSI G IG IKET+EML+ + +T IE++ D IN+AF+RL
Sbjct: 483 KPLQLPAFPLIFGKRSIKGGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLL 304
Query: 339 KNDVRYRFVVDV 350
KNDVRYRFV+D+
Sbjct: 303 KNDVRYRFVIDI 268
>BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1.1.1.1).
{Neurospora crassa}, partial (57%)
Length = 682
Score = 109 bits (272), Expect = 2e-24
Identities = 67/184 (36%), Positives = 96/184 (51%), Gaps = 6/184 (3%)
Frame = +2
Query: 29 LRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM-SNYPMVPGHEVVGEVLEVGSDVTRFTV 87
++ GPD+V + V + GVCHTD+H + D + + P+V GHE G V+ G V +
Sbjct: 146 VQKPGPDEVLVNVKFSGVCHTDLHAWQGDWPLDTKLPLVGGHEGAGVVVARGDLVKDVKI 325
Query: 88 GEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIVEQKFVV 147
GE VG L G C C+ CQ+ E C + + + G G F + I + V
Sbjct: 326 GEKVGIKWLNGSCLSCSYCQNADESLCAEAL-------LSGYTVDGSFQQYAIAKAIHVA 484
Query: 148 KIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGG----ILGL-GGVGHMGVIIAKA 202
+IPE E ++P+LCA +TVY GLKESG++ G I+G GG+G + KA
Sbjct: 485 RIPEECDLESISPILCAGITVYK-----GLKESGVKAGQSIAIVGAGGGLGSIAXQYCKA 649
Query: 203 MGHH 206
MG H
Sbjct: 650 MGIH 661
>AW126217 homologue to GP|17028158|gb| cinnamyl-alcohol dehydrogenase
{Medicago sativa}, partial (15%)
Length = 487
Score = 100 bits (250), Expect = 7e-22
Identities = 47/53 (88%), Positives = 52/53 (97%)
Frame = +1
Query: 290 LGRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDV 342
+GR+SITGSF+GS+KETEEMLEFWKEKGL+SMIEIV MDYINKAFERLEKNDV
Sbjct: 325 VGRKSITGSFVGSVKETEEMLEFWKEKGLSSMIEIVTMDYINKAFERLEKNDV 483
>TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (96%)
Length = 1524
Score = 99.0 bits (245), Expect = 3e-21
Identities = 90/371 (24%), Positives = 153/371 (40%), Gaps = 32/371 (8%)
Frame = +3
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
G R+P+ L+ F + ++ IK CGVCH+D+H +K ++ S+ P V GHE+
Sbjct: 237 GAVFREPNKPLTIEEFHIPRPKAGELLIKTKGCGVCHSDLHVMKGEIPFSS-PCVVGHEI 413
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGC----CKKCTACQSDIEQYCKKKIWNYN---DV 124
GEV+E G T+ + +VG C C+ C + C+ + YN
Sbjct: 414 TGEVVEHGQHTDSKTIERLPIGSRVVGAFIMPCGNCSYCSKGHDDLCEA-FFAYNRAKGT 590
Query: 125 YVDGKP--------------TQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYS 170
DG+ + GG AE +V + +P M + A L CA T Y
Sbjct: 591 LYDGETRLFLRGSGNPIYMYSMGGLAEYCVVPANALAVLPNSMPYTESAILGCAVFTAYG 770
Query: 171 PLSHFGLKESGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYL 230
++H G ++G GGVG + IA+A G + +K E + LGA +
Sbjct: 771 AMAHAAEVRPGDTVAVIGTGGVGSSCLQIARAFGASDIIAVDVQDEKLEKAKTLGATHTI 950
Query: 231 VSSDTTSMQEAAD-----SLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFIT 285
S+ +++ + +D ++ + +K GK +++G+
Sbjct: 951 NSAKEDPIEKILEITGGKGVDVAVEALGRPQTFAQCTQSVKDGGKAVMIGLAQAGSLGEV 1130
Query: 286 PMVMLGRRSI--TGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYI----NKAFERLEK 339
+ L RR I GS+ G ++ L E G+ + V Y KAF+ L +
Sbjct: 1131DINRLVRRKIKVIGSYGGRARQDLPKLIRLAETGIFDLGHAVTRKYTFEESGKAFQDLNE 1310
Query: 340 NDVRYRFVVDV 350
+ R V+++
Sbjct: 1311GKIVGRAVIEI 1343
>BQ158078 similar to GP|10187159|emb cDNA~Strawberry alcohol dehydrogenase
{Fragaria x ananassa}, partial (19%)
Length = 970
Score = 79.7 bits (195), Expect = 2e-15
Identities = 34/60 (56%), Positives = 43/60 (71%)
Frame = +2
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GWAARDPSG+LSP+ F+ R TG DV KV Y G+CH+D+H KN+ G YP+ GHE+
Sbjct: 26 GWAARDPSGVLSPFNFSRRETGEKDVTFKVLY*GICHSDLHMAKNEWGTYFYPLDSGHEL 205
>TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbitol
dehydrogenase {Prunus persica}, partial (93%)
Length = 1691
Score = 73.9 bits (180), Expect = 9e-14
Identities = 65/234 (27%), Positives = 109/234 (45%), Gaps = 5/234 (2%)
Frame = +2
Query: 27 FTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNY----PMVPGHEVVGEVLEVGSDV 82
F L + GP DV IK+ G+C +DVH +K L +++ PMV GHE G + EVGS V
Sbjct: 308 FNLPSLGPHDVRIKMKAVGICGSDVHYLKT-LRCADFIVKEPMVIGHECAGIIEEVGSQV 484
Query: 83 TRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIVE 142
G+ V + C +C C+ C + + P G A I+
Sbjct: 485 KTLVPGDRVAIEPGISCW-RCDHCKLGRYNLCP------DMKFFATPPVHGSLANQIVHP 643
Query: 143 QKFVVKIPEGMAPEQVAPLLCAAVTVYS-PLSHFGLKESGLRGGILGLGGVGHMGVIIAK 201
K+PE ++ E+ A +V V++ ++ G + + L I+G G +G + ++ A+
Sbjct: 644 ADLCFKLPENVSLEEGAMCEPLSVGVHACRRANIGPETNVL---IMGAGPIGLVTMLSAR 814
Query: 202 AMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPVG 255
A G V+ D + + LGAD + S T++Q+ A+ + I + + G
Sbjct: 815 AFGAPRIVVVDVDDHRLSVAKSLGADDIVKVS--TNIQDVAEEVKQIHNVLGAG 970
>TC78975 similar to GP|21592656|gb|AAM64605.1 alcohol dehydrogenase
putative {Arabidopsis thaliana}, partial (77%)
Length = 1691
Score = 64.7 bits (156), Expect = 5e-11
Identities = 83/334 (24%), Positives = 134/334 (39%), Gaps = 32/334 (9%)
Frame = +2
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSN-YPMVPGHEVVGEVLEVGSDVTRFTVGE---IV 91
+V IK+ Y +CHTD+ K + YP + GHE G V VG V + ++
Sbjct: 209 EVRIKILYTSICHTDLSGWKGECEPQRAYPRIFGHEASGIVESVGEGVNDMKENDKVVLI 388
Query: 92 GAGLLVGC----CKKCTACQSDIEQYCKKKIWNYNDVY--VDGKP-----TQGGFAETII 140
G C CKK C+ KK + + + +DGKP F E +
Sbjct: 389 FNGECGECKCCMCKKTNMCEKSGVDPMKKLMCDGTSRFSSIDGKPIFHFLNTSTFTEYTV 568
Query: 141 VEQKFVVKI--PEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGVI 198
V+ VK+ + ++ +++ L C T + +G I GLG VG
Sbjct: 569 VDSACAVKLNTEDNLSLKKLTLLSCGVSTGIGAAWNNANVHAGSSVAIFGLGAVGLAVAE 748
Query: 199 IAKAMG-HHVTVISSSDRKKKEAIEDLGADAYLVSSDTTS-----MQEAAD-SLDYIID- 250
A+A G + + + K E +G ++ D ++E D +DY +
Sbjct: 749 GARARGASKIIGVDINPDKFNNTAETMGITEFINPKDEEKPVYEIIREMTDGGVDYSFEC 928
Query: 251 TVPVGHPLEPYLSLLKLDGKLILMGVINTP-LQFITPMVMLGRRSITGSFIGSIKETEEM 309
T + + +LS+ + G +L+G+ +P L I PM + R I GS G K ++
Sbjct: 929 TGNLNVLRDSFLSVHEGWGLTVLLGIHGSPKLLPIHPMELFDGRRIEGSVFGGFKGKSQL 1108
Query: 310 LEFWKE--KGLTSMIEIVN----MDYINKAFERL 337
E KG + + D IN+AF L
Sbjct: 1109PNLATECMKGAIKLDNFITHELPFDEINQAFNLL 1210
>BQ146920 similar to GP|8099340|gb|A ELI3 {Lycopersicon esculentum}, partial
(20%)
Length = 686
Score = 63.5 bits (153), Expect = 1e-10
Identities = 30/60 (50%), Positives = 42/60 (70%)
Frame = +2
Query: 291 GRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
G R GS IG +KET++ML+F + + IE++ MDY+N A ERL K DV+YRFV+D+
Sbjct: 8 GER**PGSNIGGMKETQDMLDFAAKHNVKPTIEVIPMDYVNTAMERLLKADVKYRFVIDI 187
>TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, complete
Length = 1502
Score = 62.0 bits (149), Expect = 3e-10
Identities = 85/364 (23%), Positives = 139/364 (37%), Gaps = 30/364 (8%)
Frame = +2
Query: 8 RTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVP 67
R V W A P ++ G +V +K+ + +CHTDV+ + +P +
Sbjct: 122 RAAVAWEAGKPL-VMEEVEVAPPQAG--EVRLKILFTSLCHTDVYFWEAKGQTPLFPRIF 292
Query: 68 GHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWN------Y 121
GHE G V +G VT G+ + G C C C+S+ C N
Sbjct: 293 GHEAGGIVESIGEGVTHLKPGD-HALPVFTGECGDCPHCKSEESNMCDLLRINTDRGVMI 469
Query: 122 ND----VYVDGKPTQ-----GGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPL 172
ND + G+P F+E +V V KI ++V L C T
Sbjct: 470 NDNQSRFSLKGQPIHHFVGTSTFSEYTVVHAGCVAKINPDAPLDKVCILSCGICTGLGAT 649
Query: 173 SHFGLKESGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYL-- 230
+ + G I GLG VG A+ G + + E + G + ++
Sbjct: 650 INVAKPKPGSSVAIFGLGAVGLAAAEGARISGASRIIGVDLVSSRFELAKKFGVNEFVNP 829
Query: 231 VSSDTTSMQEAADSLDYIID-TVPVGHPLEPYLSLLKLD----GKLILMGVINTPLQFIT 285
D Q A+ + +D V ++ +S + G +L+GV N F T
Sbjct: 830 KDHDKPVQQVIAEMTNGGVDRAVECTGSIQAMISAFECVHDGWGVAVLVGVPNKDDAFKT 1009
Query: 286 -PMVMLGRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEI-------VNMDYINKAFERL 337
PM +L R++ G+F G+ K ++ EK + +E+ + INKAF+ +
Sbjct: 1010HPMNLLNERTLKGTFYGNYKPRTDLPNV-VEKYMKGELELEKFITHTIPFSEINKAFDYM 1186
Query: 338 EKND 341
K +
Sbjct: 1187LKGE 1198
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,390,566
Number of Sequences: 36976
Number of extensions: 135760
Number of successful extensions: 724
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 700
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 710
length of query: 358
length of database: 9,014,727
effective HSP length: 97
effective length of query: 261
effective length of database: 5,428,055
effective search space: 1416722355
effective search space used: 1416722355
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0290.23


