

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0290.16
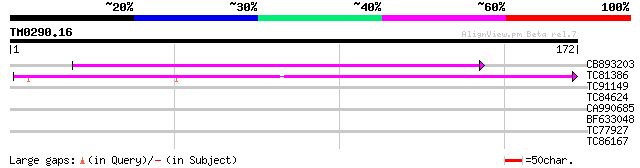
(172 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB893203 89 1e-18
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 75 1e-14
TC91149 similar to GP|15081723|gb|AAK82516.1 T10F18_70/T10F18_70... 29 1.1
TC84624 similar to GP|2462925|emb|CAA03884.1 GTP cyclohydrolase ... 28 2.4
CA990685 weakly similar to GP|22136772|gb putative carbonyl redu... 28 2.4
BF633048 similar to GP|11994760|d contains similarity to pheroph... 27 5.3
TC77927 similar to GP|21740503|emb|CAD40827. OSJNBa0006B20.18 {O... 26 9.1
TC86167 similar to PIR|H96751|H96751 probable casein kinase I F2... 26 9.1
>CB893203
Length = 800
Score = 88.6 bits (218), Expect = 1e-18
Identities = 45/125 (36%), Positives = 66/125 (52%)
Frame = -3
Query: 20 VSNEWGTFWPNCSQLVLNRDVSDHCPVLLRQTFQDWGPKPFRVLNCWLGDHRLPALVENS 79
VS W WPN Q L R +SD CP++L +WGP+ +L CW V++
Sbjct: 798 VSESWCIKWPNMIQRALFRGLSDRCPIMLTIDEGNWGPRLHHMLKCWADLPGYHLFVKDK 619
Query: 80 WKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRSRREVVVTQINNLDLKEEEVGLD 139
W +V WG +++KEK K +R+ LK+W+ +L +R + I LD K E++ LD
Sbjct: 618 WNSFQVSRWGGYVLKEK*KMVRNNLKDWHHNHTRNLDARINDIRESIAYLDSKGEDLTLD 439
Query: 140 QAEVD 144
EVD
Sbjct: 438 TEEVD 424
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 75.1 bits (183), Expect = 1e-14
Identities = 47/175 (26%), Positives = 84/175 (47%), Gaps = 4/175 (2%)
Frame = -1
Query: 2 FTW---HRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLR-QTFQDWGP 57
FTW R N + RLDR +V+ + S L + SDH P+L QT
Sbjct: 597 FTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILFELQTQNIQFS 418
Query: 58 KPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRS 117
F+ + W +++ W + +V G F++ +KLK L+ VLK WN+ FG++ S
Sbjct: 417 SSFKFMKMWSAHPDCINIIKQCWAN-QVVGCPMFVLNQKLKNLKEVLKVWNKNTFGNVHS 241
Query: 118 RREVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAILRLHESFLCQKARSKW 172
+ + +++++ +K + +G +D KA + + L + E F +K++ W
Sbjct: 240 QVDNAYKELDDIQVKIDSIGYSDVLMDQEKAAQLNLESALNIEEVFWHEKSKVNW 76
>TC91149 similar to GP|15081723|gb|AAK82516.1 T10F18_70/T10F18_70
{Arabidopsis thaliana}, partial (14%)
Length = 895
Score = 28.9 bits (63), Expect = 1.1
Identities = 16/58 (27%), Positives = 27/58 (45%)
Frame = +1
Query: 38 RDVSDHCPVLLRQTFQDWGPKPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKE 95
R ++D PVL+ + PK +++C R A ++WK+ + WG V E
Sbjct: 502 RSITDLPPVLISEILNCLDPKELGIVSCVSLILRSLASEHHAWKEFYCERWGLPAVPE 675
>TC84624 similar to GP|2462925|emb|CAA03884.1 GTP cyclohydrolase II / 3
4-dihydroxy-2-butanone-4-phoshate synthase {Arabidopsis
thaliana}, partial (37%)
Length = 628
Score = 27.7 bits (60), Expect = 2.4
Identities = 15/55 (27%), Positives = 24/55 (43%), Gaps = 9/55 (16%)
Frame = +3
Query: 4 WHRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLN---------RDVSDHCPVLLR 49
+ R ++ R L+ WG F NC + +L+ D+ D C VL+R
Sbjct: 405 YRRKRDKLVERAGAALIPTMWGPFEANCYRSLLDGMEHIAMVKGDIGDGCDVLVR 569
>CA990685 weakly similar to GP|22136772|gb putative carbonyl reductase
{Arabidopsis thaliana}, partial (36%)
Length = 554
Score = 27.7 bits (60), Expect = 2.4
Identities = 11/27 (40%), Positives = 17/27 (62%)
Frame = +2
Query: 100 LRSVLKEWNREVFGDLRSRREVVVTQI 126
L+ + EW REVFGD+ + E V ++
Sbjct: 263 LKRIKNEWTREVFGDVDNLTEEKVDEV 343
>BF633048 similar to GP|11994760|d contains similarity to
pherophorin~gene_id:T5M7.14 {Arabidopsis thaliana},
partial (15%)
Length = 510
Score = 26.6 bits (57), Expect = 5.3
Identities = 16/62 (25%), Positives = 31/62 (49%), Gaps = 3/62 (4%)
Frame = +2
Query: 94 KEKLKGLRSVLKEWNREV---FGDLRSRREVVVTQINNLDLKEEEVGLDQAEVDLRKALF 150
K L+ R+ +KE R++ + + ++ Q++ L +KEE + AE+D +
Sbjct: 62 KRDLELARNKIKELQRQMQLEANQTKGQLLLLKQQVSGLQVKEEAGAIKDAEIDKKLKAV 241
Query: 151 ND 152
ND
Sbjct: 242 ND 247
>TC77927 similar to GP|21740503|emb|CAD40827. OSJNBa0006B20.18 {Oryza
sativa}, partial (17%)
Length = 867
Score = 25.8 bits (55), Expect = 9.1
Identities = 10/25 (40%), Positives = 16/25 (64%)
Frame = -2
Query: 83 MKVKGWGAFMVKEKLKGLRSVLKEW 107
M+ W A ++EKL G+ V+K+W
Sbjct: 137 MQWNHWIAVFLQEKLHGMECVVKKW 63
>TC86167 similar to PIR|H96751|H96751 probable casein kinase I F28P22.10
[imported] - Arabidopsis thaliana, partial (74%)
Length = 1967
Score = 25.8 bits (55), Expect = 9.1
Identities = 12/27 (44%), Positives = 15/27 (55%)
Frame = -2
Query: 30 NCSQLVLNRDVSDHCPVLLRQTFQDWG 56
N LVL + +HC +LLRQT G
Sbjct: 508 NPRDLVLVNPLQEHCTLLLRQTISHSG 428
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,491,071
Number of Sequences: 36976
Number of extensions: 90685
Number of successful extensions: 405
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 404
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 404
length of query: 172
length of database: 9,014,727
effective HSP length: 89
effective length of query: 83
effective length of database: 5,723,863
effective search space: 475080629
effective search space used: 475080629
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0290.16


