

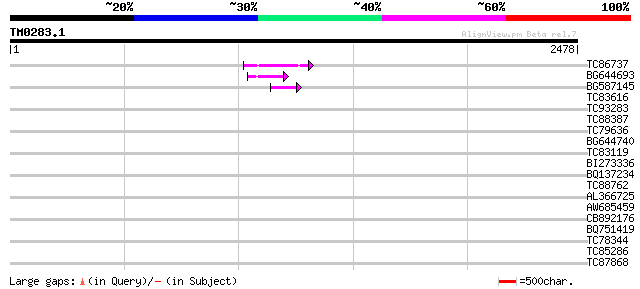
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0283.1
(2478 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 132 2e-30
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 99 2e-20
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 67 9e-11
TC83616 similar to GP|16226661|gb|AAL16226.1 At2g28910/F8N16.20 ... 39 0.033
TC93283 weakly similar to GP|23476966|emb|CAD43403. SMC3 protein... 37 0.096
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 34 0.81
TC79636 similar to PIR|T00587|T00587 probable ubiquitin--protein... 33 1.1
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 33 1.1
TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 33 1.4
BI273336 homologue to PIR|T09415|T09 probable surface protein - ... 33 1.4
BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkho... 33 1.4
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 32 2.4
AL366725 32 3.1
AW685459 32 3.1
CB892176 PIR|T09286|T09 malate dehydrogenase (EC 1.1.1.37) precu... 32 4.0
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 32 4.0
TC78344 similar to GP|13540405|gb|AAK29456.1 histone H1 {Lens cu... 31 5.2
TC85286 similar to GP|3776572|gb|AAC64889.1| ESTs gb|R65052 gb|... 31 5.2
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 31 5.2
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 132 bits (332), Expect = 2e-30
Identities = 86/314 (27%), Positives = 159/314 (50%), Gaps = 7/314 (2%)
Frame = +1
Query: 1020 ALPYIDEFNEKDIPTKARPIQ-MNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLN 1078
A+P I + + D P P+ M+ Q L ++ + + L+KG I+ S S +V
Sbjct: 421 AIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKTLEDLLDKGFIKASGSAAGAPVLFVRK 600
Query: 1079 ASELERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVR 1138
G +R ++Y+ LN + K RYPLP ++ ++R+ F+K D+ + ++++ ++
Sbjct: 601 PG----GGIRFCVDYRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIK 768
Query: 1139 EEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYM-NFTIVYLDDVLVFSK 1197
+ED+ KTAF +G +EW V P GL AP+ +Q +N + ++ +F Y+DDVL+++
Sbjct: 769 DEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTT 948
Query: 1198 GIDQ-HVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQ---ITPIQRSLEF 1253
G + H + + + + GL + KK + T +++G+ + G+ P++ +
Sbjct: 949 GSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIR 1128
Query: 1254 AKKFPDELKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVRE 1312
P +K + FLG NY DFIP + PL + RK+ P W E + +
Sbjct: 1129 DWLPPGSVK---GARSFLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFTK 1299
Query: 1313 VKKLVQSLPIVTPF 1326
+K+L P++ F
Sbjct: 1300 LKRLFAEEPVLRMF 1341
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 99.4 bits (246), Expect = 2e-20
Identities = 61/180 (33%), Positives = 98/180 (53%), Gaps = 1/180 (0%)
Frame = +2
Query: 1040 QMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYKPLNKV 1099
++N LK + ++ + L KG I+ S P + ++ + + G LR+ I+Y LN V
Sbjct: 92 RINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVLFL----KKKDGFLRMSIDYPQLNNV 259
Query: 1100 LKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVM 1159
I+YPLP +L L F K D++ G +Q V ED KTAF + +GHYE VM
Sbjct: 260 NIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPKTAFRIRYGHYEILVM 439
Query: 1160 PQGLKNAPSEYQNIMNDIFYPYM-NFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGL 1218
G N P + +MN +F Y+ + IV+ +D+L++SK ++H HL ++++K GL
Sbjct: 440 SFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEHENHLRLALKVLKDIGL 619
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 67.0 bits (162), Expect = 9e-11
Identities = 44/134 (32%), Positives = 68/134 (49%), Gaps = 1/134 (0%)
Frame = +2
Query: 1140 EDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKG 1198
+D KTAF+ G Y + VMP GLKNA S YQ ++N +F + T+ VY+DD+LV S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 1199 IDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFP 1258
H+ HL++ + + + + ++ K T FLGY + Q I + + P
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 1259 DELKEKTQLQRFLG 1272
K ++QR G
Sbjct: 371 SP-KNSREVQRLTG 409
>TC83616 similar to GP|16226661|gb|AAL16226.1 At2g28910/F8N16.20
{Arabidopsis thaliana}, partial (30%)
Length = 854
Score = 38.5 bits (88), Expect = 0.033
Identities = 30/93 (32%), Positives = 43/93 (45%), Gaps = 8/93 (8%)
Frame = +2
Query: 671 KNVCWKCGKPGHYANKCKTQQKINELDLDQKLKDSLISVLI--------NSEDPHYSSEY 722
+ C KCG+ GH +CK KI + D ++K D++ S+ + +E S
Sbjct: 392 RGACKKCGRVGHLKFQCKNNVKIKD-DKEEKDFDAMQSLGLVRMDKLKGKAEKVDKGSNV 568
Query: 723 EYEGDEDQDINLVEYSSDSHSSSEEERECVKDS 755
E +E+ D SSDS SE ER VK S
Sbjct: 569 ESSEEEEDD----SESSDSEIDSEIERAIVKRS 655
>TC93283 weakly similar to GP|23476966|emb|CAD43403. SMC3 protein
{Arabidopsis thaliana}, partial (18%)
Length = 902
Score = 37.0 bits (84), Expect = 0.096
Identities = 42/166 (25%), Positives = 68/166 (40%), Gaps = 20/166 (12%)
Frame = +1
Query: 722 YEYEGDEDQDINLVEYSSDSHSSSEEERECVKDSSGFCDCAGCLGQNIN-MLTQDQTMSL 780
Y++ + +N ++ ++DS EEE E VK D Q IN ++T+ Q +
Sbjct: 352 YDHRRSRLKFMNTIKQNADSIHVKEEELEKVKFEIQEID------QKINDLVTEQQKVDA 513
Query: 781 ISIIDKMEDSPLRDEFLQQ-------LNLLVKKEETLKKDISPPINSMN----------- 822
K E L+ + L KKE++L+ D+ I +
Sbjct: 514 QCAHSKSEIEELKRDIANSNKQKPLFSKALAKKEKSLE-DVQNQIEQLKGSIATKKAEMG 690
Query: 823 -EIFNRFRPKPTITLSDLQEEVKILKEEINQLKQNDLSLEFRLIEL 867
E+ + P LSDL E+K LKE++ K + + E R EL
Sbjct: 691 TELIDHLTPXEKKLLSDLNPEIKDLKEKLVACKTDRIESEARKAEL 828
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 33.9 bits (76), Expect = 0.81
Identities = 24/107 (22%), Positives = 48/107 (44%), Gaps = 5/107 (4%)
Frame = +1
Query: 625 SKARRINRSKTKPQGYKKRKPFQKKPYEQNPSTQQPTNKKNVNP--KKKNVCWKCGKPGH 682
S++R +RS++ + R P ++ + S ++ +++ + N+C C +PGH
Sbjct: 271 SRSRSKSRSRSPMDRSRSRSPVDRRIRSERFSHREAPYRRDSRRGFSQDNLCKNCKRPGH 450
Query: 683 YANKCKTQQKINELDLDQKLKD--SLISVLINSEDP-HYSSEYEYEG 726
Y +C + L + S S+ N ++P H +S EG
Sbjct: 451 YVRECPNVAVCHNCSLPGHIASECSTKSLCWNCKEPGHMASSCPNEG 591
>TC79636 similar to PIR|T00587|T00587 probable ubiquitin--protein ligase (EC
6.3.2.19) - Arabidopsis thaliana, partial (18%)
Length = 876
Score = 33.5 bits (75), Expect = 1.1
Identities = 18/36 (50%), Positives = 26/36 (72%)
Frame = +2
Query: 2166 ISSILGIPSSVLKKLLSSLSLFKTSNLKLVPLKLRL 2201
IS +L +P SVL+KLLSS+ +L+L+ LKL+L
Sbjct: 629 ISRLLFLPKSVLRKLLSSMITATVISLQLLLLKLKL 736
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 33.5 bits (75), Expect = 1.1
Identities = 15/47 (31%), Positives = 25/47 (52%)
Frame = -1
Query: 1085 GALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSG 1131
G R+ I+Y+ NKV +YPLP +L ++ + F D++ G
Sbjct: 208 GYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NIDLRLG 68
>TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 421
Score = 33.1 bits (74), Expect = 1.4
Identities = 18/67 (26%), Positives = 33/67 (48%), Gaps = 4/67 (5%)
Frame = +3
Query: 625 SKARRINRSKTKPQGYKK----RKPFQKKPYEQNPSTQQPTNKKNVNPKKKNVCWKCGKP 680
S++R +RS+++ +K R ++ PY ++ S + N+C C +P
Sbjct: 216 SRSRSRSRSRSRSPRIRKIRSDRHSYRDAPYRRDSSR---------GFSRDNLCKNCKRP 368
Query: 681 GHYANKC 687
GHYA +C
Sbjct: 369 GHYAREC 389
>BI273336 homologue to PIR|T09415|T09 probable surface protein - alfalfa,
partial (71%)
Length = 589
Score = 33.1 bits (74), Expect = 1.4
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 1/65 (1%)
Frame = +1
Query: 1368 PLKSAGSSAPATPSKSSQRLTTSQIVKTPPQNQKAP-LVPLANKYTVLTQSSSKSPVKPE 1426
P+ G+SAPATP+ S+ T + + + P VP + V T ++S S V E
Sbjct: 322 PIVKVGASAPATPATLSKPATPAAVSTSSAGEVATPAAVPASGAGQVTTPAASPSVVIAE 501
Query: 1427 STEYF 1431
S E F
Sbjct: 502 SPESF 516
>BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkholderia
cepacia}, partial (6%)
Length = 1184
Score = 33.1 bits (74), Expect = 1.4
Identities = 15/46 (32%), Positives = 26/46 (55%)
Frame = +1
Query: 626 KARRINRSKTKPQGYKKRKPFQKKPYEQNPSTQQPTNKKNVNPKKK 671
+ARR R+K + Q K+RK ++KP + + T KK P+++
Sbjct: 238 EARRAGRNKRRSQRQKERKENERKPKTKQKKGGEETRKKKKQPQRR 375
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 32.3 bits (72), Expect = 2.4
Identities = 21/88 (23%), Positives = 38/88 (42%)
Frame = +3
Query: 1335 YRVNMSEKPKSKDQY*PPLGSSSTSKDKALAITPLKSAGSSAPATPSKSSQRLTTSQIVK 1394
Y+ N P S P+ ++++ A +P+ + S +TP+ S TTS
Sbjct: 69 YQFNNPTSPSSSPSASSPVSTTTSPPASTPASSPVSTTTSPPASTPASSPVPTTTSPPAP 248
Query: 1395 TPPQNQKAPLVPLANKYTVLTQSSSKSP 1422
TP + + P A+ L +++ SP
Sbjct: 249 TPASSPVSTNSPTASPAGSLPAAATPSP 332
>AL366725
Length = 485
Score = 32.0 bits (71), Expect = 3.1
Identities = 20/71 (28%), Positives = 32/71 (44%)
Frame = +2
Query: 618 EPLQAPSSKARRINRSKTKPQGYKKRKPFQKKPYEQNPSTQQPTNKKNVNPKKKNVCWKC 677
+P AP+ K ++ +P+ KK P + + + +K NV PK+ C +C
Sbjct: 218 KPYSAPADKGKQRMVDDRRPK--KKDAPAEIVCFNYG----EKGHKSNVCPKEIKKCVRC 379
Query: 678 GKPGHYANKCK 688
K GH CK
Sbjct: 380 DKKGHIVADCK 412
>AW685459
Length = 632
Score = 32.0 bits (71), Expect = 3.1
Identities = 29/101 (28%), Positives = 47/101 (45%), Gaps = 6/101 (5%)
Frame = -3
Query: 2308 RLIIKYKIPS-AIIKNGSLEIETPFLLVRNLSQKVIIGTP---FIKKLFPYDTDVNGITV 2363
RLI + KI S A I IE PF + S +++ P F +FP+D+ +
Sbjct: 594 RLIYRKKI*SFAYIIELKRRIELPFFFLGKQSTSLLVLLPTVFFQPHIFPFDSQCLQLGS 415
Query: 2364 QHLGQPILFKFSEPPFDKTLNVI--SYKEKQINFLKEEISY 2402
H+ + F +TLN++ S + K +KE+I+Y
Sbjct: 414 DHIKRREFLSRKSITFVETLNILVQSERGKLSITVKEDIAY 292
>CB892176 PIR|T09286|T09 malate dehydrogenase (EC 1.1.1.37) precursor -
alfalfa, partial (41%)
Length = 615
Score = 31.6 bits (70), Expect = 4.0
Identities = 26/63 (41%), Positives = 33/63 (52%)
Frame = -2
Query: 1840 HHRAEEKASGFHLPFSLSLSLSLSLSFP*VCL*FLKFLFE*KLVSVSTLSFHIFIFAKCF 1899
HH EKA L SLSLSLSLSLS P V F F F+ +S + F+ +K
Sbjct: 197 HHGCFEKALSLSLSLSLSLSLSLSLS-PRVFF-FFFFFFDITKLSCK*NAITTFLSSKIK 24
Query: 1900 LSS 1902
+S+
Sbjct: 23 MSN 15
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 31.6 bits (70), Expect = 4.0
Identities = 23/76 (30%), Positives = 38/76 (49%), Gaps = 1/76 (1%)
Frame = +1
Query: 1351 PPLGSSSTSKDKALAITPLKSAGSSAP-ATPSKSSQRLTTSQIVKTPPQNQKAPLVPLAN 1409
P SSTS+ ++A S+P ATP+++ TS +TPP + + P A+
Sbjct: 361 PSASRSSTSRPPRARAR--RAASRSSPWATPARACTTAPTSASRQTPPPSPRTAAKPKAS 534
Query: 1410 KYTVLTQSSSKSPVKP 1425
+ T S S++P +P
Sbjct: 535 R---CTPSRSRAPTEP 573
>TC78344 similar to GP|13540405|gb|AAK29456.1 histone H1 {Lens culinaris},
partial (86%)
Length = 1215
Score = 31.2 bits (69), Expect = 5.2
Identities = 20/82 (24%), Positives = 31/82 (37%)
Frame = +2
Query: 1342 KPKSKDQY*PPLGSSSTSKDKALAITPLKSAGSSAPATPSKSSQRLTTSQIVKTPPQNQK 1401
KPK K + P+ + +K KA A A + A A P+K+ K P +
Sbjct: 506 KPKPKPKAKAPVAKAPAAKSKAKAAPAKAKAKAKAKAAPAKAKPAAKAKPAAKAKPAAKA 685
Query: 1402 APLVPLANKYTVLTQSSSKSPV 1423
P+ K ++ PV
Sbjct: 686 KPVAKAKPKAKAPVAKANAVPV 751
>TC85286 similar to GP|3776572|gb|AAC64889.1| ESTs gb|R65052 gb|AA712146
gb|H76533 gb|H76282 gb|AA650771 gb|H76287 gb|AA650887
gb|N37383, partial (77%)
Length = 1947
Score = 31.2 bits (69), Expect = 5.2
Identities = 36/132 (27%), Positives = 58/132 (43%), Gaps = 5/132 (3%)
Frame = +2
Query: 1841 HRAEEKASGFHLPFSLSLSLSLSLSFP*VCL*FLKFLFE*KLVSVSTLSFHIFIFAKCFL 1900
+RA + L SLSLSLSLSLS L L +LS + + L
Sbjct: 680 NRAPPLSLSLSLSLSLSLSLSLSLS--------LSLSLSLSLSLSLSLSLSLSLSLSLSL 835
Query: 1901 SSAPSGKRDSW-----QAFLNLLATKFGKNLEAKILGEKSHIKLLAMVSS*LLFKELPLK 1955
S P+ R ++ + + L+ T F N + +L KS L +S + +K + +
Sbjct: 836 SLVPNSARGTFPHSNPPSLIFLMTTLFPPNSLSPLLNLKSSPSLQPKSTSLISYKPI-VS 1012
Query: 1956 SVKMTKSKSNSP 1967
++K T++ S P
Sbjct: 1013 NLKKTQTLSLKP 1048
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 31.2 bits (69), Expect = 5.2
Identities = 8/17 (47%), Positives = 15/17 (88%)
Frame = +3
Query: 674 CWKCGKPGHYANKCKTQ 690
C++CG+PGH+A +C+ +
Sbjct: 336 CYECGEPGHFARECRNR 386
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 76,866,291
Number of Sequences: 36976
Number of extensions: 1156308
Number of successful extensions: 8074
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 6379
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7691
length of query: 2478
length of database: 9,014,727
effective HSP length: 112
effective length of query: 2366
effective length of database: 4,873,415
effective search space: 11530499890
effective search space used: 11530499890
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0283.1


