

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
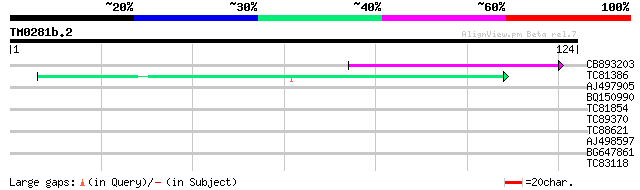
Query= TM0281b.2
(124 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB893203 57 1e-09
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 40 2e-04
AJ497905 similar to SP|Q26636|CATL_ Cathepsin L precursor (EC 3.... 28 1.2
BQ150990 28 1.2
TC81854 weakly similar to PIR|T50012|T50012 hypothetical protein... 27 2.0
TC89370 weakly similar to PIR|T01856|T01856 hypothetical protein... 26 4.5
TC88621 similar to PIR|H84698|H84698 hypothetical protein At2g29... 26 4.5
AJ498597 similar to GP|666056|emb| conglutin gamma {Lupinus angu... 25 5.9
BG647861 similar to GP|6815055|dbj EST AU081527(C53732) correspo... 25 7.7
TC83118 similar to PIR|E96718|E96718 probable cullin T6C23.13 [i... 25 7.7
>CB893203
Length = 800
Score = 57.4 bits (137), Expect = 1e-09
Identities = 21/47 (44%), Positives = 28/47 (58%)
Frame = -3
Query: 75 LSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+S WC WP+ +Q + R LSD CP+M + NWGPR +L CW
Sbjct: 798 VSESWCIKWPNMIQRALFRGLSDRCPIMLTIDEGNWGPRLHHMLKCW 658
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 40.4 bits (93), Expect = 2e-04
Identities = 28/106 (26%), Positives = 41/106 (38%), Gaps = 3/106 (2%)
Frame = -1
Query: 7 FCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWF---RPN 63
+C GDFN + S E G R M DF+ + +L P G +TW R
Sbjct: 741 WCCIGDFNTILGSHEHQGSHTP--ARLPMLDFQQWSDVNNLFHLPTRGSAFTWTNGRRGR 568
Query: 64 GQAASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMN 109
RLDR +++ + + SDH P++F N
Sbjct: 567 NNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILFELQTQN 430
>AJ497905 similar to SP|Q26636|CATL_ Cathepsin L precursor (EC 3.4.22.15).
[Flesh fly Boettcherisca peregrina] {Sarcophaga
peregrina}, partial (26%)
Length = 306
Score = 27.7 bits (60), Expect = 1.2
Identities = 9/31 (29%), Positives = 17/31 (54%)
Frame = +1
Query: 71 DRFLLSSDWCSCWPHGVQTVMARSLSDHCPV 101
D +++ + WC+ W MAR+ +HC +
Sbjct: 214 DYWIVKNSWCTSWGQDGFIKMARNRDNHCGI 306
>BQ150990
Length = 974
Score = 27.7 bits (60), Expect = 1.2
Identities = 15/43 (34%), Positives = 23/43 (52%)
Frame = -1
Query: 37 DFKAFLSNLSLIEPPAVGKKYTWFRPNGQAASRLDRFLLSSDW 79
DF+ L+ +++ E PA + WF P G+ +R D S DW
Sbjct: 221 DFRV-LAYINISEVPASDETRLWFLPEGRG*ARGDYGSHSGDW 96
>TC81854 weakly similar to PIR|T50012|T50012 hypothetical protein T31P16.70
- Arabidopsis thaliana, partial (29%)
Length = 1124
Score = 26.9 bits (58), Expect = 2.0
Identities = 9/26 (34%), Positives = 15/26 (57%)
Frame = -2
Query: 76 SSDWCSCWPHGVQTVMARSLSDHCPV 101
S+ W +C P G ++ R L+ CP+
Sbjct: 754 SNSWSNCMPFGSLLLICRQLT*RCPI 677
>TC89370 weakly similar to PIR|T01856|T01856 hypothetical protein F9D12.19 -
Arabidopsis thaliana, partial (15%)
Length = 710
Score = 25.8 bits (55), Expect = 4.5
Identities = 11/28 (39%), Positives = 15/28 (53%), Gaps = 1/28 (3%)
Frame = +3
Query: 98 HCPVMFRSTDMNWGPRPFRVLNCW-FSD 124
HCP + T WG + ++ CW FSD
Sbjct: 459 HCPPGWHFTLGIWGQQSIKLAQCWSFSD 542
>TC88621 similar to PIR|H84698|H84698 hypothetical protein At2g29650
[imported] - Arabidopsis thaliana, partial (49%)
Length = 1074
Score = 25.8 bits (55), Expect = 4.5
Identities = 14/36 (38%), Positives = 16/36 (43%), Gaps = 4/36 (11%)
Frame = -1
Query: 54 GKKYTWFRPNGQAASRLDRFL----LSSDWCSCWPH 85
G +Y WFR N L RFL S+ WPH
Sbjct: 1005 GPRYRWFRQN*SPRYFLHRFLGETICSNQLLFFWPH 898
>AJ498597 similar to GP|666056|emb| conglutin gamma {Lupinus angustifolius},
partial (42%)
Length = 707
Score = 25.4 bits (54), Expect = 5.9
Identities = 8/14 (57%), Positives = 10/14 (71%)
Frame = -1
Query: 75 LSSDWCSCWPHGVQ 88
L +C CWP+GVQ
Sbjct: 206 LKGFFCGCWPNGVQ 165
>BG647861 similar to GP|6815055|dbj EST AU081527(C53732) corresponds to a
region of the predicted gene.~hypothetical protein,
partial (72%)
Length = 733
Score = 25.0 bits (53), Expect = 7.7
Identities = 10/31 (32%), Positives = 16/31 (51%)
Frame = +2
Query: 78 DWCSCWPHGVQTVMARSLSDHCPVMFRSTDM 108
DW CW H + +S +CP + + TD+
Sbjct: 143 DWFCCWWHYKCILWIQSCYANCP*ICKRTDV 235
>TC83118 similar to PIR|E96718|E96718 probable cullin T6C23.13 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 902
Score = 25.0 bits (53), Expect = 7.7
Identities = 15/48 (31%), Positives = 20/48 (41%)
Frame = -3
Query: 37 DFKAFLSNLSLIEPPAVGKKYTWFRPNGQAASRLDRFLLSSDWCSCWP 84
+FKAF +LSL+ + Y F + F LS SC P
Sbjct: 210 EFKAFWKDLSLLNAWFIILSYLSFSSKSLCTNSTGSFNLSGSVTSCLP 67
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.137 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,983,262
Number of Sequences: 36976
Number of extensions: 65155
Number of successful extensions: 473
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 472
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 473
length of query: 124
length of database: 9,014,727
effective HSP length: 84
effective length of query: 40
effective length of database: 5,908,743
effective search space: 236349720
effective search space used: 236349720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0281b.2


