

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0281a.3
(1430 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
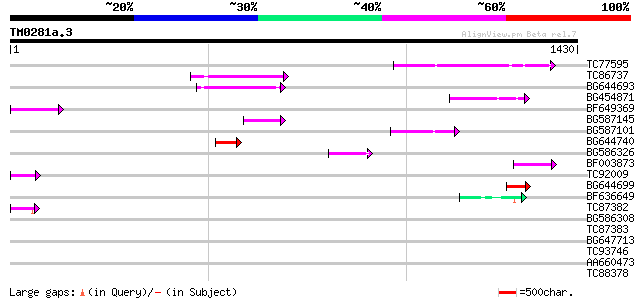
Score E
Sequences producing significant alignments: (bits) Value
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 201 1e-51
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 176 4e-44
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 160 3e-39
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 135 9e-32
BF649369 96 1e-19
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 89 1e-17
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 73 9e-13
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 67 4e-11
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 64 4e-10
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 61 3e-09
TC92009 56 9e-08
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 51 4e-06
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 49 1e-05
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 45 3e-04
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 41 0.004
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 40 0.006
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 40 0.006
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 35 0.16
AA660473 35 0.21
TC88378 similar to GP|18086484|gb|AAL57695.1 At1g07280/F22G5_32 ... 34 0.47
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 201 bits (512), Expect = 1e-51
Identities = 137/424 (32%), Positives = 217/424 (50%), Gaps = 16/424 (3%)
Frame = +2
Query: 968 IISECHDTPTGGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLL 1027
++ E HD+ GH G T + ++ +W G ++ FV +C++C A G L
Sbjct: 179 LVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGFL 358
Query: 1028 QPLPIPTQVWMDISMDFVGGLPKVRGKDT--VLVVVDRLTKFAHFYALGHPYSAKDVAVL 1085
+PLP+P ++ D+SMDF+ LP RG+ + + V+VDRL+K + A+ A
Sbjct: 359 KPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMD-TMEAEACAQR 535
Query: 1086 FIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCL 1145
F+ + HG P +IVSDR ++ FW+E ++ G LSTSYHPQTDG TE N+ +
Sbjct: 536 FLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEI 715
Query: 1146 ETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGR--DP-PLLLKAGAIP 1202
+ LR +V W D LP + +NSS G +PF +G DP P + G +
Sbjct: 716 QAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYHVDPIPTVEDTGGVV 895
Query: 1203 SRVEEVNQLFQQR-DGVLGELKQNLLKAQHQMKVQADKHRRLVE-FNEGDWVYLKLQPYK 1260
S E QL +R V G ++ ++ AQ + + A+K R + + GD V+L + YK
Sbjct: 896 SEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEASANKRRCPADRYQVGDKVWLNVSNYK 1075
Query: 1261 LKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELPAHSKIHPVFHVSLLKQALK---PT 1317
+ RP KL + Y++ V P +L +P ++P FHV LL++A P
Sbjct: 1076 ----SPRPSKKL-DWLHHKYEVTRFVTPHVVELNVP--GTVYPKFHVDLLRRAASDPLPG 1234
Query: 1318 QQ---PQPLPPMLNEELELEVVPEDVIDHREDAQG---NLEVLIKWEQLPTCDNTWELAA 1371
Q+ PQP PP+++++ E+E E+++ R G + L+KW+ D TWE A
Sbjct: 1235 QEVVDPQP-PPIVDDDGEVEWEVEEILAARWHQVGRGRRRQALVKWKGF--VDATWEAAD 1405
Query: 1372 VIQD 1375
I++
Sbjct: 1406 AIRE 1417
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 176 bits (447), Expect = 4e-44
Identities = 104/261 (39%), Positives = 156/261 (58%), Gaps = 13/261 (4%)
Frame = +1
Query: 456 GVLLQFDTVFQDPQGLP---PRRRHDHAIHL---KEGAQVPNLRPYKCPHYQKNEVEKLV 509
GVL +F +F + R DHAI L K+G P P+ P Y + E LV
Sbjct: 340 GVLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPP--LPWG-PLYGMSRQELLV 510
Query: 510 A-----EMLEAGVIRPSISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEE 564
++L+ G I+ S S +P++ V+K GG R CVDYRALN +T +++P+P+I E
Sbjct: 511 LKKTLEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISE 690
Query: 565 LLDELGGAKVFSKLDLKSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQS 624
L + GA+ F+KLD+ + +H++R+KD D EKTAFRT G +E++V PFGLT AP+TFQ
Sbjct: 691 TLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQR 870
Query: 625 LMNEVLKPYLRKFALVFFDDILVYSP-DLLTHTTHLQQVLTLLADHALKANRKKCSFGQT 683
+N+ L +L F + DD+L+Y+ H +++VL LAD L + KKC F T
Sbjct: 871 YINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVT 1050
Query: 684 SLEYLGHIV-SGEGGGC*PIK 703
+++Y+G I+ +G+G C P+K
Sbjct: 1051TVKYVGFILTAGKGVSCDPLK 1113
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 160 bits (406), Expect = 3e-39
Identities = 97/236 (41%), Positives = 131/236 (55%), Gaps = 10/236 (4%)
Frame = +2
Query: 471 LPPRRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVA-----EMLEAGVIRPSISPF 525
+PP + D I L +PN+ P P Y+ N ++ V ++LE G I+PSI P
Sbjct: 17 VPPEWKIDFGIDL-----LPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP* 181
Query: 526 SSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYH 585
++ +KKKDG R+ +DY LN V I K+P+P+I+EL D L G+K F K+DL+ G H
Sbjct: 182 GVVVLFLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*H 361
Query: 586 QIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDI 645
Q R+ D+ KTAFR GHYE LVM FG TN P F LMN V + YL +VF +DI
Sbjct: 362 QHRVIGEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDI 541
Query: 646 LVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQT-SLEYLG----HIVSGEG 696
L+YS + H HL+ L +L D L C L +G H++SGEG
Sbjct: 542 LIYSKNENEHENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEG 691
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 135 bits (341), Expect = 9e-32
Identities = 75/202 (37%), Positives = 111/202 (54%)
Frame = +2
Query: 1110 SNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEF 1169
SNFWK+LFK+ GT L +S++YHP +DGQ+E N+ E YLRC + P +W PWAE+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1170 WFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKA 1229
W+NT+YN SA M+PFKALYGRD +L+++ ++ QR+ +L +L+ +
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQREELLSQLQSISTRL 391
Query: 1230 QHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPV 1289
K+ + K R WV L L + C + + ++
Sbjct: 392 N---KL*SIKLIRNAAILSSSWVSTSL*NCNLINSLR*HCGNIKSSVHP--TLVHY*QSA 556
Query: 1290 AYKLELPAHSKIHPVFHVSLLK 1311
AYKL LP+ +K+ P+FHVS LK
Sbjct: 557 AYKLSLPSTAKVPPIFHVSQLK 622
>BF649369
Length = 631
Score = 95.9 bits (237), Expect = 1e-19
Identities = 43/135 (31%), Positives = 79/135 (57%)
Frame = +3
Query: 1 RKLEIPIFSGDDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWERCNPSP 60
+K+++P+F GDD V W + E YF ++ +D R++ + +++EG + WF
Sbjct: 120 KKVKLPLFEGDDPVAWITRAEIYFDVQNTPDDMRVKLSRLSMEGPTIHWFNLLMETEDDL 299
Query: 61 TWEAFKLAVVRRFQPSMIQNPFELLLSLKQVGTVDEYVEEFEKYAGALKEIDHDFVRGIF 120
+ E K A++ R+ ++NPFE L +L+Q+G+V+E+VE FE + + + + G F
Sbjct: 300 SREKLKKALIARYDGRRLENPFEELSTLRQIGSVEEFVEAFELLSSQVGRLPEEQYLGYF 479
Query: 121 LNGLKEEIKAEVRLI 135
++GLK I+ VR +
Sbjct: 480 MSGLKAHIRRRVRTL 524
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 89.0 bits (219), Expect = 1e-17
Identities = 48/108 (44%), Positives = 65/108 (59%)
Frame = +2
Query: 589 MKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVY 648
M D+EKTAF T G Y + VMPFGL NA ST+Q L+N + L V+ DD+LV
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 649 SPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEG 696
S H HL++ L ++ +K N KC+FG TS E+LG+IV+ +G
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQG 325
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 72.8 bits (177), Expect = 9e-13
Identities = 48/175 (27%), Positives = 82/175 (46%), Gaps = 2/175 (1%)
Frame = +2
Query: 961 ASNRIPMIISECHDTPTGGHSGHFRTYKRIASF-LYWEGMKTDIKNFVEDCEICQRSKYS 1019
A IP I+ CH + GH +T +I +W M D +F+ C+ CQR
Sbjct: 83 AEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNI 262
Query: 1020 TLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSA 1079
+ + Q + +V+ +DF+G P +LV VD ++K+ A+ P +
Sbjct: 263 S*R-NEMPQNFILEVEVFDVWGIDFMGPFPSSYNNKYILVAVDYVSKWVE--AIASPTND 433
Query: 1080 KDVAVLFIKELV-KLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQ 1133
V V K ++ G P ++SD F++ +++L K G + K++T+YHPQ
Sbjct: 434 ATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 67.4 bits (163), Expect = 4e-11
Identities = 30/65 (46%), Positives = 45/65 (69%)
Frame = -1
Query: 519 RPSISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKL 578
+PSISP + ++ V+KKDG +R+C+DYR NKVT NK+P+P I+ L D++ F +
Sbjct: 262 QPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NI 83
Query: 579 DLKSG 583
DL+ G
Sbjct: 82 DLRLG 68
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 63.9 bits (154), Expect = 4e-10
Identities = 38/111 (34%), Positives = 59/111 (52%)
Frame = +2
Query: 804 QTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQRL 863
+ +A+ S+ L + ++ E+ A+V A++ WR YL G + TD KSLK++ Q
Sbjct: 107 KVIAYASRQLRKHEGNYPTHDLEMAAVVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPE 286
Query: 864 FS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSALSILTSAEKE 914
+ Q +W + D +I Y PG N VADALSR+ + SAE+E
Sbjct: 287 LNLRQRRWMEFVADYDLDITYYPGKANLVADALSRR------RVDVSAERE 421
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 61.2 bits (147), Expect = 3e-09
Identities = 38/113 (33%), Positives = 60/113 (52%), Gaps = 5/113 (4%)
Frame = +2
Query: 1271 KLAPRFYGPYKILDRVGPVAYKLELPAH-SKIHPVFHVSLLKQAL-KPTQQPQPLPPMLN 1328
KL RF GPY+I +RVG VAY++ LP H +H VFHVS L++ + P+ Q +
Sbjct: 29 KLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHVIQSDDVQVR 208
Query: 1329 EELELEVVPEDVIDHREDAQGNLE---VLIKWEQLPTCDNTWELAAVIQDKFP 1378
+ L +E +P + D + E V + W++ TWEL + + + +P
Sbjct: 209 DNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVESYP 367
>TC92009
Length = 974
Score = 56.2 bits (134), Expect = 9e-08
Identities = 25/77 (32%), Positives = 44/77 (56%), Gaps = 1/77 (1%)
Frame = +2
Query: 2 KLEIPIFSGDDAVGWTQKLERYFAMREVTEDERMQATMVALEG-RALSWFQWWERCNPSP 60
K ++ F+G D GW + E +FA E+ +++Q +++E +AL WF W + NP
Sbjct: 680 KYKLSQFTGTDPAGWITRAEMFFADNEIHSCDKLQWAFMSMEDEKALLWFYSWCQENPDA 859
Query: 61 TWEAFKLAVVRRFQPSM 77
W++F +A++R F M
Sbjct: 860 DWKSFSMAMIREFGARM 910
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 50.8 bits (120), Expect = 4e-06
Identities = 26/62 (41%), Positives = 39/62 (61%), Gaps = 1/62 (1%)
Frame = +2
Query: 1252 VYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLEL-PAHSKIHPVFHVSLL 1310
V LK+ P + KL+ R+ GP++++ R+G VAY+L L P S +HPVFHVS+
Sbjct: 8 VLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVSMF 187
Query: 1311 KQ 1312
K+
Sbjct: 188 KR 193
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 49.3 bits (116), Expect = 1e-05
Identities = 46/186 (24%), Positives = 74/186 (39%), Gaps = 17/186 (9%)
Frame = +1
Query: 1134 TDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPP 1193
+D Q + N LET+L F + +L WAE +NTN++ +AG +PFK +Y
Sbjct: 7 SDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY----- 171
Query: 1194 LLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVY 1253
A + L + +G+ K F G Y
Sbjct: 172 ----VVAHLQKFVVARDLIYRNEGL-------------------HKSST*TSFGRGTRAY 282
Query: 1254 --LKLQPYKLKSIATRPCA-----------KLAPRF----YGPYKILDRVGPVAYKLELP 1296
L Y+ RP + ++ P++ YGPY+++ ++G VA+KL LP
Sbjct: 283 EALSRPAYETC*HPRRPLSIVYTRDRTYEWQVLPKYVA*CYGPYQVIKQIGSVAFKL*LP 462
Query: 1297 AHSKIH 1302
+IH
Sbjct: 463 EQHQIH 480
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 44.7 bits (104), Expect = 3e-04
Identities = 24/84 (28%), Positives = 44/84 (51%), Gaps = 10/84 (11%)
Frame = +2
Query: 2 KLEIPIFSG----DDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWER-- 55
K++IP F G DD + W Q +ER F +EV E+++++ L+ AL W++ +R
Sbjct: 617 KVDIPDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRRR 796
Query: 56 ----CNPSPTWEAFKLAVVRRFQP 75
+ TW+ + + R++ P
Sbjct: 797 KREGKSKIKTWDKMRQKLTRKYLP 868
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 40.8 bits (94), Expect = 0.004
Identities = 22/71 (30%), Positives = 38/71 (52%)
Frame = -2
Query: 1094 HGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFV 1153
HG P IV+D F+SN ++E + +L ++ +PQ++GQ E +N+ + L+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 1154 GPKPKQWLDWL 1164
K W D L
Sbjct: 505 DLKKGCWADEL 473
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 40.0 bits (92), Expect = 0.006
Identities = 24/84 (28%), Positives = 42/84 (49%), Gaps = 10/84 (11%)
Frame = -2
Query: 2 KLEIPIFSG----DDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWER-- 55
K +IP F G DD + W Q +ER F +EV E+++++ L+ A W++ +R
Sbjct: 736 K*DIPDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRR 557
Query: 56 ----CNPSPTWEAFKLAVVRRFQP 75
+ TWE + + R++ P
Sbjct: 556 KREGKSKIKTWEKMRQKLTRKYLP 485
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 40.0 bits (92), Expect = 0.006
Identities = 26/101 (25%), Positives = 52/101 (50%), Gaps = 11/101 (10%)
Frame = +2
Query: 2 KLEIPIFSG----DDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWER-- 55
K++IP F G D+ V W Q +ER F +EV E+++++ L+ A W++ +R
Sbjct: 335 KVDIPDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKNLKRKR 514
Query: 56 -C---NPSPTWEAFKLAVVRRF-QPSMIQNPFELLLSLKQV 91
C + TW+ + + R++ P Q+ F +++++
Sbjct: 515 NCEGKSKIKTWDKMRQKLTRKYLHPHYYQDNFTQKKNIQKI 637
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 35.4 bits (80), Expect = 0.16
Identities = 23/74 (31%), Positives = 40/74 (53%), Gaps = 1/74 (1%)
Frame = +2
Query: 998 GMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDF-VGGLPKVRGKDT 1056
G D++ F + ++C LL+P+P P W D+++DF +G L + KD+
Sbjct: 416 GFLADMEPFKDKTKLC------------LLRPVPKPP--WEDVTIDFSLGLL*TQQLKDS 553
Query: 1057 VLVVVDRLTKFAHF 1070
+VV D+ ++ AHF
Sbjct: 554 KMVVGDKFSRMAHF 595
>AA660473
Length = 655
Score = 35.0 bits (79), Expect = 0.21
Identities = 41/152 (26%), Positives = 61/152 (39%), Gaps = 26/152 (17%)
Frame = -2
Query: 260 GEFEDALSGEFNSLQLSLCSMAGLTSTKSWK-----MVGQIGNQSIVVLIDCGASHNFIS 314
GE ED L +NS+ LC M K K +VG + IV+L+D GA+HNF
Sbjct: 630 GEKEDILV--YNSM--GLCEMTEFNKLKVVKPATLQLVGCLKGVPIVILVDIGANHNF-- 469
Query: 315 HKLVEQLNLTVAETQTYTVEVGDGYKIRCKGKC-----SQLGIMIQALEI---------V 360
+ V D + CKG+ S++G + + + V
Sbjct: 468 ---------------DFASSVSD---VGCKGRVILSQESEVG*WSENINVG*AY*SRGTV 343
Query: 361 QNFYLFG-------LNGMDVVLGIEWLASLGD 385
+ F+ G L D+ LG+ WL LG+
Sbjct: 342 RGFHC*GVDAYELELGEFDMFLGVAWLEKLGN 247
>TC88378 similar to GP|18086484|gb|AAL57695.1 At1g07280/F22G5_32 {Arabidopsis
thaliana}, partial (38%)
Length = 1687
Score = 33.9 bits (76), Expect = 0.47
Identities = 15/45 (33%), Positives = 25/45 (55%)
Frame = +2
Query: 168 SKVVTVEPQNNTDQKQDSSSVGSVAMPSRNNAGIDSSRNRGGEFK 212
S +V V N+ +K DSS + + ++ N A +D RGG++K
Sbjct: 1067 SSIVEVHNDNHVFKKFDSSIIKTFSVNGNNTASVDGGSGRGGKYK 1201
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,365,851
Number of Sequences: 36976
Number of extensions: 692230
Number of successful extensions: 2924
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 2875
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2916
length of query: 1430
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1322
effective length of database: 5,021,319
effective search space: 6638183718
effective search space used: 6638183718
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0281a.3


