

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0280a.5
(151 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
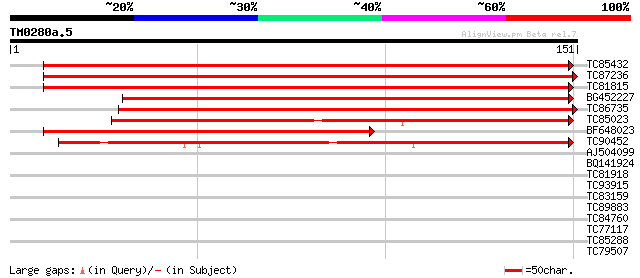
Score E
Sequences producing significant alignments: (bits) Value
TC85432 homologue to GP|7381203|gb|AAF61434.1| pathogenesis-rela... 232 4e-62
TC87236 homologue to GP|7381203|gb|AAF61434.1| pathogenesis-rela... 223 2e-59
TC81815 similar to GP|7381203|gb|AAF61434.1| pathogenesis-relate... 216 2e-57
BG452227 homologue to GP|7381203|gb| pathogenesis-related protei... 216 4e-57
TC86735 similar to GP|6562381|emb|CAB62537.1 pseudo-hevein {Heve... 184 9e-48
TC85023 weakly similar to GP|7381205|gb|AAF61435.1| pre-hevein-l... 144 1e-35
BF648023 similar to GP|7381203|gb|A pathogenesis-related protein... 143 2e-35
TC90452 weakly similar to GP|19879970|gb|AAM00217.1 class 4 path... 125 5e-30
AJ504099 36 0.005
BQ141924 similar to PIR|F86387|F863 probable Pto kinase interact... 29 0.82
TC81918 similar to GP|21594031|gb|AAM65949.1 unknown {Arabidopsi... 29 0.82
TC93915 similar to GP|10178125|dbj|BAB11537. gene_id:MOP10.2~unk... 29 0.82
TC83159 27 4.1
TC89883 homologue to PIR|C86303|C86303 hypothetical protein AAG0... 27 4.1
TC84760 similar to GP|8567777|gb|AAF76349.1| unknown protein {Ar... 27 4.1
TC77117 similar to GP|9294182|dbj|BAB02084.1 protein kinases-lik... 27 4.1
TC85288 homologue to SP|Q43070|GAE1_PEA UDP-glucose 4-epimerase ... 26 6.9
TC79507 similar to GP|14194103|gb|AAK56246.1 AT3g22390/MCB17_12 ... 25 9.1
>TC85432 homologue to GP|7381203|gb|AAF61434.1| pathogenesis-related protein
4A {Pisum sativum}, partial (96%)
Length = 614
Score = 232 bits (592), Expect = 4e-62
Identities = 104/141 (73%), Positives = 118/141 (82%)
Frame = +2
Query: 10 TQRKLSIVAVWWLIAATFANGQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLA 69
TQR LS++ + +L+ NGQSANNV ATY+LYNPQ I WN TA YCATW+AN+PL+
Sbjct: 29 TQRTLSLLVLCFLVGTMLVNGQSANNVRATYHLYNPQNINWNYNTASVYCATWDANQPLS 208
Query: 70 WRKKYGWTAFCGPAGAHGRDSCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRL 129
WR+KYGWTAFCGP G HGRDSCGKCLRV NTATG +ATVRIVDQCANGGLDLDVNVF ++
Sbjct: 209 WRQKYGWTAFCGPQGPHGRDSCGKCLRVTNTATGAQATVRIVDQCANGGLDLDVNVFNQI 388
Query: 130 DSNGQGNHQGHLTVNYRFVNC 150
D+NGQG QGHLTVNY FVNC
Sbjct: 389 DTNGQGYQQGHLTVNYVFVNC 451
>TC87236 homologue to GP|7381203|gb|AAF61434.1| pathogenesis-related protein
4A {Pisum sativum}, partial (93%)
Length = 701
Score = 223 bits (569), Expect = 2e-59
Identities = 100/142 (70%), Positives = 115/142 (80%)
Frame = +2
Query: 10 TQRKLSIVAVWWLIAATFANGQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLA 69
TQRKLS++ + +L+ +GQSANNV ATYN YNPQ I W+ TA YCATW+AN+PL+
Sbjct: 8 TQRKLSLLVLCFLVGTMLVSGQSANNVRATYNNYNPQNINWDYNTASVYCATWDANQPLS 187
Query: 70 WRKKYGWTAFCGPAGAHGRDSCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRL 129
WR KYGWTAFCGPAG GRDSCGKCL V NTATG + TVRIVDQC+NGGLDLDVNVF ++
Sbjct: 188 WRSKYGWTAFCGPAGPTGRDSCGKCLTVTNTATGAQVTVRIVDQCSNGGLDLDVNVFNQI 367
Query: 130 DSNGQGNHQGHLTVNYRFVNCG 151
D+NGQG GHLTVNY FVNCG
Sbjct: 368 DTNGQGVQNGHLTVNYSFVNCG 433
>TC81815 similar to GP|7381203|gb|AAF61434.1| pathogenesis-related protein
4A {Pisum sativum}, partial (84%)
Length = 578
Score = 216 bits (551), Expect = 2e-57
Identities = 97/141 (68%), Positives = 114/141 (80%)
Frame = +1
Query: 10 TQRKLSIVAVWWLIAATFANGQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLA 69
TQRKLS++ +L+ A+ QSAN V +TY+LYNPQ I W+ A YCATW+AN+PL
Sbjct: 37 TQRKLSLLVFCFLVGMMLASNQSANGVRSTYHLYNPQNINWDYNRASVYCATWDANQPLE 216
Query: 70 WRKKYGWTAFCGPAGAHGRDSCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRL 129
WRKKYGWTAFCGP G GRDSCGKCLRV+NTATG + TVRIVDQCANGGLDLDVNVF+R+
Sbjct: 217 WRKKYGWTAFCGPQGPRGRDSCGKCLRVKNTATGAQETVRIVDQCANGGLDLDVNVFKRI 396
Query: 130 DSNGQGNHQGHLTVNYRFVNC 150
D+NGQG +GHL V+Y FVNC
Sbjct: 397 DTNGQGYQKGHLIVDYVFVNC 459
>BG452227 homologue to GP|7381203|gb| pathogenesis-related protein 4A {Pisum
sativum}, partial (81%)
Length = 677
Score = 216 bits (549), Expect = 4e-57
Identities = 96/120 (80%), Positives = 105/120 (87%)
Frame = +3
Query: 31 QSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLAWRKKYGWTAFCGPAGAHGRDS 90
QSANNV ATY+LYNPQ I WN TA YCATW+AN+PL+WR+KYGWTAFCGP G HGRDS
Sbjct: 3 QSANNVRATYHLYNPQNINWNYNTASVYCATWDANQPLSWRQKYGWTAFCGPQGPHGRDS 182
Query: 91 CGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRLDSNGQGNHQGHLTVNYRFVNC 150
CGKCLRV NTATG +ATVRIVDQCANGGLDLDVNVF ++D+NGQG QGHLTVNY FVNC
Sbjct: 183 CGKCLRVTNTATGAQATVRIVDQCANGGLDLDVNVFNQIDTNGQGYQQGHLTVNYVFVNC 362
>TC86735 similar to GP|6562381|emb|CAB62537.1 pseudo-hevein {Hevea
brasiliensis}, partial (83%)
Length = 863
Score = 184 bits (468), Expect = 9e-48
Identities = 79/122 (64%), Positives = 98/122 (79%)
Frame = +2
Query: 30 GQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLAWRKKYGWTAFCGPAGAHGRD 89
G+SA+NV ATY+ Y P Q GW+L AYC+TW+A+KP +WR KYGWTAFCGP G G+
Sbjct: 221 GESASNVRATYHYYQPDQHGWDLNAVSAYCSTWDASKPYSWRSKYGWTAFCGPVGPRGQA 400
Query: 90 SCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRLDSNGQGNHQGHLTVNYRFVN 149
SCGKCLRV N+ TG + TVRIVDQC+NGGLDLDV VF R+D++G+G QGHL V+Y+FV+
Sbjct: 401 SCGKCLRVTNSGTGAQETVRIVDQCSNGGLDLDVGVFNRIDTDGRGYQQGHLIVSYQFVD 580
Query: 150 CG 151
CG
Sbjct: 581 CG 586
>TC85023 weakly similar to GP|7381205|gb|AAF61435.1| pre-hevein-like protein
{Pisum sativum}, partial (42%)
Length = 550
Score = 144 bits (364), Expect = 1e-35
Identities = 70/125 (56%), Positives = 86/125 (68%), Gaps = 2/125 (1%)
Frame = +3
Query: 28 ANGQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLAWRKKYGWTAFCGPAGAHG 87
A+ QSANNV ATY YNP +I WNL T+G +CA + NK L+WR ++GW AFCG A G
Sbjct: 129 ASAQSANNVKATYQSYNPHKIKWNLNTSGVFCAAQDGNKSLSWRSEFGWAAFCG--HAVG 302
Query: 88 RDSCGKCLRVRNTATG--DEATVRIVDQCANGGLDLDVNVFRRLDSNGQGNHQGHLTVNY 145
+ CGKCL V N G VRIVD C NGGL LD++VF++LDS G GN QG+L V+Y
Sbjct: 303 KGVCGKCLNVTNKENGIIVSKIVRIVDTCTNGGLKLDIDVFQKLDSFGTGNAQGYLMVDY 482
Query: 146 RFVNC 150
FV+C
Sbjct: 483 EFVDC 497
>BF648023 similar to GP|7381203|gb|A pathogenesis-related protein 4A {Pisum
sativum}, partial (60%)
Length = 482
Score = 143 bits (361), Expect = 2e-35
Identities = 61/88 (69%), Positives = 71/88 (80%)
Frame = +2
Query: 10 TQRKLSIVAVWWLIAATFANGQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLA 69
TQR LS++ + +L+ NGQSANNV ATY+LYNPQ I WN TA YCATW+AN+PL+
Sbjct: 20 TQRTLSLLVLCFLVGTMLVNGQSANNVRATYHLYNPQNINWNYNTASVYCATWDANQPLS 199
Query: 70 WRKKYGWTAFCGPAGAHGRDSCGKCLRV 97
WR+KYGWTAFCGP G HGRDSCGKCLRV
Sbjct: 200 WRQKYGWTAFCGPQGPHGRDSCGKCLRV 283
>TC90452 weakly similar to GP|19879970|gb|AAM00217.1 class 4
pathogenesis-related protein {Prunus persica}, partial
(79%)
Length = 720
Score = 125 bits (315), Expect = 5e-30
Identities = 70/152 (46%), Positives = 95/152 (62%), Gaps = 15/152 (9%)
Frame = +3
Query: 14 LSIVAVWWLIAATFANGQSANNVMATYNLYNP----QQIG--WNLLTAGAYCATWNANKP 67
L +V ++ ++ A+ QS NV ATY +N Q IG WNL TAG CA+ + NK
Sbjct: 36 LDLVLIFCVLV--LASAQSEINVNATYRSFNSPTTKQGIGNKWNLNTAGVLCASQDGNKS 209
Query: 68 LAWRKKYGWTAFCGPAGAHGRDSCGKCLRVRNTATGDEA---------TVRIVDQCANGG 118
L+WR KYGW AFCG + + +CGKCL V N +G TVRIVD C++GG
Sbjct: 210 LSWRSKYGWAAFCGIWLS--KYACGKCLNVTNRDSGSTVAQLRFPITQTVRIVDNCSHGG 383
Query: 119 LDLDVNVFRRLDSNGQGNHQGHLTVNYRFVNC 150
L+LD++VF++LD++G G QG+L V+Y FVNC
Sbjct: 384 LELDMDVFQKLDTSGYGMSQGYLVVDYEFVNC 479
>AJ504099
Length = 610
Score = 36.2 bits (82), Expect = 0.005
Identities = 21/64 (32%), Positives = 31/64 (47%)
Frame = +1
Query: 87 GRDSCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRLDSNGQGNHQGHLTVNYR 146
G D+C KCL + N + V+I+DQCA G V++ R S G + + R
Sbjct: 421 GLDNCYKCLEITNPKNKKKVVVKIIDQCA-GCKKNQVDLTRTAFSKIANPDDGIVHIKSR 597
Query: 147 FVNC 150
V+C
Sbjct: 598 VVSC 609
>BQ141924 similar to PIR|F86387|F863 probable Pto kinase interactor
[imported] - Arabidopsis thaliana, partial (6%)
Length = 1314
Score = 28.9 bits (63), Expect = 0.82
Identities = 22/58 (37%), Positives = 24/58 (40%), Gaps = 6/58 (10%)
Frame = +3
Query: 80 CGPAGAHG----RDSCGKCLRVRNTATGDEATVRIVDQCANGGLDL--DVNVFRRLDS 131
CG GAH R G C G TVR D C GG+ + V FR LDS
Sbjct: 387 CGAGGAHSSAGRRSGGGGCGGGEEVDRGRIRTVRATDGCTAGGVWVVDRVFAFRGLDS 560
>TC81918 similar to GP|21594031|gb|AAM65949.1 unknown {Arabidopsis
thaliana}, partial (78%)
Length = 1051
Score = 28.9 bits (63), Expect = 0.82
Identities = 9/17 (52%), Positives = 11/17 (63%)
Frame = +3
Query: 80 CGPAGAHGRDSCGKCLR 96
CGP GAHG C C++
Sbjct: 696 CGPEGAHG*SQCSSCIK 746
>TC93915 similar to GP|10178125|dbj|BAB11537. gene_id:MOP10.2~unknown
protein {Arabidopsis thaliana}, partial (29%)
Length = 1001
Score = 28.9 bits (63), Expect = 0.82
Identities = 12/26 (46%), Positives = 15/26 (57%)
Frame = -3
Query: 70 WRKKYGWTAFCGPAGAHGRDSCGKCL 95
WR GW +CG G G DS G+C+
Sbjct: 327 WRAMEGWGVWCGKRG--GVDSVGECV 256
>TC83159
Length = 371
Score = 26.6 bits (57), Expect = 4.1
Identities = 12/27 (44%), Positives = 15/27 (55%)
Frame = +1
Query: 47 QIGWNLLTAGAYCATWNANKPLAWRKK 73
Q W L YC WN KPLA+++K
Sbjct: 277 QCAWVLTWLEIYC--WNIGKPLAYQQK 351
>TC89883 homologue to PIR|C86303|C86303 hypothetical protein AAG09080.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1287
Score = 26.6 bits (57), Expect = 4.1
Identities = 8/24 (33%), Positives = 13/24 (53%)
Frame = +3
Query: 49 GWNLLTAGAYCATWNANKPLAWRK 72
GWNLL + + W ++P W +
Sbjct: 93 GWNLLVFQSLSSVWRLSQPTGWAR 164
>TC84760 similar to GP|8567777|gb|AAF76349.1| unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 781
Score = 26.6 bits (57), Expect = 4.1
Identities = 11/32 (34%), Positives = 14/32 (43%)
Frame = +1
Query: 49 GWNLLTAGAYCATWNANKPLAWRKKYGWTAFC 80
GW L G CA + ++ W K WT C
Sbjct: 634 GWTLSATGGVCAVXSVSQK*HWASK--WTTVC 723
>TC77117 similar to GP|9294182|dbj|BAB02084.1 protein kinases-like protein
{Arabidopsis thaliana}, partial (34%)
Length = 955
Score = 26.6 bits (57), Expect = 4.1
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = -2
Query: 19 VWWLIAATFANGQSAN 34
VWW+IAA+F GQ N
Sbjct: 48 VWWVIAASFLVGQEFN 1
>TC85288 homologue to SP|Q43070|GAE1_PEA UDP-glucose 4-epimerase (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase). [Garden pea], partial (78%)
Length = 1272
Score = 25.8 bits (55), Expect = 6.9
Identities = 10/23 (43%), Positives = 12/23 (51%)
Frame = +1
Query: 58 YCATWNANKPLAWRKKYGWTAFC 80
Y +T A K L W+ KYG C
Sbjct: 847 YASTDKAQKELGWKAKYGVEEMC 915
>TC79507 similar to GP|14194103|gb|AAK56246.1 AT3g22390/MCB17_12
{Arabidopsis thaliana}, partial (11%)
Length = 2117
Score = 25.4 bits (54), Expect = 9.1
Identities = 11/22 (50%), Positives = 16/22 (72%), Gaps = 2/22 (9%)
Frame = +2
Query: 118 GLDLDVNVFR--RLDSNGQGNH 137
G+DL + + + R+DSNG GNH
Sbjct: 128 GIDLQLELEKTDRVDSNGNGNH 193
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.134 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,619,695
Number of Sequences: 36976
Number of extensions: 82937
Number of successful extensions: 325
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 320
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 321
length of query: 151
length of database: 9,014,727
effective HSP length: 88
effective length of query: 63
effective length of database: 5,760,839
effective search space: 362932857
effective search space used: 362932857
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0280a.5


