

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0278.2
(507 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
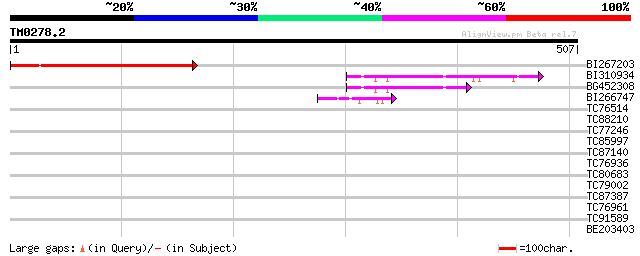
Score E
Sequences producing significant alignments: (bits) Value
BI267203 206 2e-53
BI310934 similar to PIR|T06700|T06 hypothetical protein T29H11.6... 83 2e-16
BG452308 similar to PIR|T06700|T06 hypothetical protein T29H11.6... 68 7e-12
BI266747 similar to PIR|T05497|T054 hypothetical protein T19K4.1... 42 7e-04
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 36 0.031
TC88210 similar to GP|12322899|gb|AAG51437.1 putative cell divis... 31 1.3
TC77246 similar to PIR|C71408|C71408 probable protein kinase - A... 30 2.2
TC85997 similar to PIR|JN0505|JN0505 protein kinase (EC 2.7.1.37... 30 2.9
TC87140 similar to GP|16648734|gb|AAL25559.1 AT4g23890/T32A16_60... 30 2.9
TC76936 similar to GP|20160782|dbj|BAB89723. putative nascent po... 29 3.7
TC80683 homologue to GP|8777424|dbj|BAA97014.1 gb|AAF56406.1~gen... 29 3.7
TC79002 similar to GP|15375305|gb|AAK54745.2 putative transcript... 29 4.9
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 29 4.9
TC76961 similar to GP|1841870|gb|AAB68395.1| elongation factor 1... 28 6.4
TC91589 similar to GP|5531416|emb|CAB50925.1 translocon Tic40 {P... 28 8.3
BE203403 similar to GP|12322785|gb unknown protein; 24439-25635 ... 28 8.3
>BI267203
Length = 619
Score = 206 bits (523), Expect = 2e-53
Identities = 112/169 (66%), Positives = 130/169 (76%), Gaps = 1/169 (0%)
Frame = +3
Query: 1 MGSQRLFSNSSQPMLPFVEEKKDMKLNKGLEASPGSKKHVIKGNDANSYSIEIERLRNDP 60
MGS+R FSNSSQP+ FVE K DM L KGLEAS GSK H +K ND + SIEIERLRNDP
Sbjct: 114 MGSKRPFSNSSQPISSFVEVKNDMNL-KGLEASSGSKNHTVKENDVDGCSIEIERLRNDP 290
Query: 61 SIVDTMTVQELRKTLKSIRVPAKGRKEDLLSTLKSFMDNNMGEQHPQIEEEHGLLISSEN 120
+IVDTMTV ELRKTLKSIRVPAKGRKEDLLS LK+FMDNN+ EQ QI +E GL ISSEN
Sbjct: 291 AIVDTMTVPELRKTLKSIRVPAKGRKEDLLSALKNFMDNNISEQASQIRDEQGLFISSEN 470
Query: 121 TSVEVKTKKVD-EDHVDDINDNPEVFEHSRGKRRLKQSGSESETVKVTT 168
TS+E+ +KV E+ V +++D E ++GKRRLKQS ES+ K TT
Sbjct: 471 TSLEMNAEKVSGEEPVGEVDDTLXTVELNQGKRRLKQSXPESKXXKATT 617
>BI310934 similar to PIR|T06700|T06 hypothetical protein T29H11.60 -
Arabidopsis thaliana, partial (30%)
Length = 620
Score = 83.2 bits (204), Expect = 2e-16
Identities = 64/199 (32%), Positives = 98/199 (49%), Gaps = 23/199 (11%)
Frame = +2
Query: 302 GYENSFWTCSVSKLGYSGTAIISR--IKPLSVRYGLG--ISDHDTEGRLVTAEFDTFYLI 357
G + +W+ + SK Y+GTA+ + KP SV + L S H+ +GR++ EF+T L+
Sbjct: 35 GNYHVWWSLAESK--YAGTALFVKKCFKPKSVVFNLDKIASKHEPDGRVILVEFETLRLL 208
Query: 358 CGYIPNSGDGLKRLSY-RVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNP--- 413
Y+PN+G + S+ R +WD + ++ + KP++ GDLN +HEEID+ P
Sbjct: 209 NTYVPNNGWKEEPNSFQRRRKWDKRILEFVNQ-NSDKPLIWCGDLNVSHEEIDVSQPEFF 385
Query: 414 ---------AGNKK---SAGFTDEERKSFAANFLSRGFVDTFRRQHPD---VVGYTYWGY 458
NK+ GFT ER F VD +R H D G++ W
Sbjct: 386 STAKLNGYVPPNKEDYGQPGFTLAERARFGTILREGKLVDAYRFLHKDKDMEQGFS-WSG 562
Query: 459 RHGGRKTNRGWRLDYFLVS 477
GR + R+DYF+VS
Sbjct: 563 NPIGRYRGKRMRIDYFIVS 619
>BG452308 similar to PIR|T06700|T06 hypothetical protein T29H11.60 -
Arabidopsis thaliana, partial (22%)
Length = 678
Score = 68.2 bits (165), Expect = 7e-12
Identities = 41/117 (35%), Positives = 68/117 (58%), Gaps = 5/117 (4%)
Frame = +3
Query: 302 GYENSFWTCSVSKLGYSGTAIISR--IKPLSVRYGLG--ISDHDTEGRLVTAEFDTFYLI 357
G + +W+ + SK Y+GTA+ + KP SV + L S H+ +GR++ EF+T L+
Sbjct: 69 GNYHVWWSLAESK--YAGTALFVKKCFKPKSVVFNLDKIASKHEPDGRVILVEFETLRLL 242
Query: 358 CGYIPNSGDGLKRLSY-RVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNP 413
Y+PN+G + S+ R +WD + ++ + KP++ GDLN +HEEID+ P
Sbjct: 243 NTYVPNNGWKEEPNSFQRRRKWDKRILEFVNQ-NSDKPLIWCGDLNVSHEEIDVSQP 410
>BI266747 similar to PIR|T05497|T054 hypothetical protein T19K4.180 -
Arabidopsis thaliana, partial (14%)
Length = 433
Score = 41.6 bits (96), Expect = 7e-04
Identities = 30/88 (34%), Positives = 44/88 (49%), Gaps = 17/88 (19%)
Frame = +2
Query: 276 DFDVLCLQETKLQEKDINEIKRQLIDGYENSFWTCS----VSKLGYSGTAIISRIK---- 327
D D+LC QETKL +D+ ++DGYE SF++C+ + GYSG R++
Sbjct: 137 DADILCFQETKLSRQDVT-ADLVMVDGYE-SFFSCTRTSQKGRTGYSGVITFCRVRSAFS 310
Query: 328 ------PLSVR---YGLGISDHDTEGRL 346
PL+ GL S +EG+L
Sbjct: 311 SNEAALPLAAEEGFTGLLCSSQTSEGKL 394
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 36.2 bits (82), Expect = 0.031
Identities = 43/177 (24%), Positives = 69/177 (38%), Gaps = 3/177 (1%)
Frame = +3
Query: 33 SPGSKKHVIKGNDANSY-SIEIERLRNDPSIVDTMTVQE-LRKTLKSIRVPAKGRKEDLL 90
+P K KGN + E +D S D V++ + K + S A +K D
Sbjct: 348 APSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAKKVDTS 527
Query: 91 STLKSFMDNNMGEQHPQIEEEHGLLISSENTSVEVKTKKVDEDHVDDINDNPEVFEHSRG 150
+ + E + ++ + S+N S K DE+ D+ +D E E
Sbjct: 528 E------EEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPAA 689
Query: 151 KRRLKQSGSESETVKVTTKKKLLVESDEVSDFKPS-RAKRRVSSDIVRVVSQSDEIS 206
K ++GS T ES + D KP+ +A + VS+ + S SDE S
Sbjct: 690 KAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDEES 860
>TC88210 similar to GP|12322899|gb|AAG51437.1 putative cell division related
protein; 50012-47994 {Arabidopsis thaliana}, partial
(33%)
Length = 928
Score = 30.8 bits (68), Expect = 1.3
Identities = 43/196 (21%), Positives = 76/196 (37%), Gaps = 22/196 (11%)
Frame = +3
Query: 50 SIEIERLRNDPSIVDTMTVQELRKTLKSIRVPAKGRKEDLLSTLKSFMDNNMGEQHPQIE 109
S +IE+LR + + M +E+ + ++++R D LS K+ +D +Q+ ++
Sbjct: 378 SFDIEQLRG---LCEKMEGKEVSEQVEALR--------DALSCKKNVVDEKSNQQNGSVK 524
Query: 110 EEHGLLISSENTSVEVKTKKVDEDHVDDINDNPEVFEHSRGKR-----RLKQSGSESETV 164
SS +E K K ++ ++ + + F +R +G E +
Sbjct: 525 ANGSS--SSLAGYIEKKEKPWTKEEIELLRKGIQKFPKGTSRRWEVVSEYIGTGRSVEEI 698
Query: 165 KVTTKKKLLVESDEVSDFKPSRAKRRVSSDIVRV----------VS-------QSDEIST 207
TK LL + D F KR+ ++ + VS SD +T
Sbjct: 699 MKATKTVLLQKPDTAKAFDTFLEKRKPAAQTIASPLSTREELEGVSIPATKH*NSDAKTT 878
Query: 208 TTIPTEPWTVLAHKKP 223
TTIPT T P
Sbjct: 879 TTIPTPTMTTATISTP 926
>TC77246 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (65%)
Length = 1715
Score = 30.0 bits (66), Expect = 2.2
Identities = 16/39 (41%), Positives = 25/39 (64%)
Frame = -1
Query: 43 GNDANSYSIEIERLRNDPSIVDTMTVQELRKTLKSIRVP 81
GN ++SIE E LR + ++ DT+TV E + K ++VP
Sbjct: 512 GNLEITFSIEEEILRFEIAVSDTVTVAETKSGNKLLKVP 396
>TC85997 similar to PIR|JN0505|JN0505 protein kinase (EC 2.7.1.37) 5 -
Arabidopsis thaliana, partial (78%)
Length = 2199
Score = 29.6 bits (65), Expect = 2.9
Identities = 33/163 (20%), Positives = 61/163 (37%), Gaps = 12/163 (7%)
Frame = +1
Query: 45 DANSYSIEIERLRNDPSIVDTMTVQELRKTLKSIRVPAKGRKEDLLSTLKSFMDNNMGEQ 104
D+ S + + + S + T ++L+ + S++ K EDL ++++ N+GE
Sbjct: 199 DSKSSARSLSKQHQKTSGIQTPEAKDLKFSSLSLQKTGKSEPEDLSKSVQNTSKQNIGEI 378
Query: 105 H-------PQIEEEHGLLISSE-----NTSVEVKTKKVDEDHVDDINDNPEVFEHSRGKR 152
+ P + +SS+ ++SV E +D E
Sbjct: 379 YESKKFGVPSQKGSSADSLSSKFGSGASSSVVNVNTGSQESSIDQDKKTSEYGSVKNSSV 558
Query: 153 RLKQSGSESETVKVTTKKKLLVESDEVSDFKPSRAKRRVSSDI 195
K S S K + K+ +D V K S + SSD+
Sbjct: 559 SAKASDGASSIAKTSGSAKISDRADFVESGKSSICRGSTSSDV 687
>TC87140 similar to GP|16648734|gb|AAL25559.1 AT4g23890/T32A16_60
{Arabidopsis thaliana}, partial (69%)
Length = 916
Score = 29.6 bits (65), Expect = 2.9
Identities = 13/46 (28%), Positives = 24/46 (51%)
Frame = +1
Query: 346 LVTAEFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEK 391
+V + + FY+ CG + DG + + WD ++ L+ELE+
Sbjct: 637 IVKNQNNPFYMYCGIVQRITDGKAGVLFEGGNWDKLITFRLEELER 774
>TC76936 similar to GP|20160782|dbj|BAB89723. putative nascent polypeptide
associated complex alpha chain {Oryza sativa (japonica
cultivar-group)}, partial (86%)
Length = 914
Score = 29.3 bits (64), Expect = 3.7
Identities = 25/82 (30%), Positives = 38/82 (45%)
Frame = +3
Query: 131 DEDHVDDINDNPEVFEHSRGKRRLKQSGSESETVKVTTKKKLLVESDEVSDFKPSRAKRR 190
DED D+ +DN E FE R KQ+ SE ++ K K + + SR +
Sbjct: 183 DEDDDDEDDDNAEGFE-GDASGRSKQTRSEKKSRKAMLKLGMKAVTG------VSRVTIK 341
Query: 191 VSSDIVRVVSQSDEISTTTIPT 212
S +I+ V+S+ D + T T
Sbjct: 342 KSKNILFVISKPDVFKSPTSDT 407
>TC80683 homologue to GP|8777424|dbj|BAA97014.1
gb|AAF56406.1~gene_id:K9P8.7~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (16%)
Length = 1360
Score = 29.3 bits (64), Expect = 3.7
Identities = 38/160 (23%), Positives = 63/160 (38%), Gaps = 15/160 (9%)
Frame = +1
Query: 18 VEEKKDMKLNKGLEASPGSKKHVIKGNDANSYSIEIERLRNDPSIVDTMTVQELRKTLKS 77
VEE+ D + K + + GN + I E L D S + + KT S
Sbjct: 304 VEEQSDSESEKNVADGETAADSERNGNLSADSPIPSEDLLADTSQTSSAAINA--KTTVS 477
Query: 78 IRVPAKGRK-EDLLST--LKSFMDNNMGEQHPQIEE--EHGLLISS----------ENTS 122
AK +++L + L F N + PQ+EE + L + S ENT
Sbjct: 478 DDCSAKDPSTKNMLDSEKLSDFSGNGLASVSPQLEEILDRALGLGSVAKSNKSYEAENTQ 657
Query: 123 VEVKTKKVDEDHVDDINDNPEVFEHSRGKRRLKQSGSESE 162
+++ ++ +E + D P + + R R+LK E
Sbjct: 658 LDLSSENHNESSKPAVRDKPYISKAER--RKLKNEPKHGE 771
>TC79002 similar to GP|15375305|gb|AAK54745.2 putative transcription factor
MYB124 {Arabidopsis thaliana}, partial (54%)
Length = 2304
Score = 28.9 bits (63), Expect = 4.9
Identities = 34/139 (24%), Positives = 65/139 (46%), Gaps = 23/139 (16%)
Frame = +3
Query: 57 RNDPSIVDTMTVQELRKTLKSIRVPAKGRKEDLLST---LKSFMDNNM-----GEQHP-- 106
++DP I M EL +L +++V A+ + L + L+ FM+ N G + P
Sbjct: 1215 KDDPKINALMQQAELLTSL-ALKVDAENMDQSLENAWKILQEFMNRNKESDVSGYKIPDL 1391
Query: 107 QIEEEHGLLISSENTSVEVKTKKVDEDHVDDINDNPEVFEHSRGKRRL------------ 154
Q+ + LL +N S E++ +++ D+PE EHS G L
Sbjct: 1392 QLVDLKDLLQDLKNNSEEIEPCW---RYMEFYEDSPESSEHSTGSTTLPHCTGENLEHSL 1562
Query: 155 -KQSGSESETVKVTTKKKL 172
+ SG+E +++++ +K++
Sbjct: 1563 HQDSGTELKSIQIEHQKEI 1619
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 28.9 bits (63), Expect = 4.9
Identities = 21/76 (27%), Positives = 37/76 (48%), Gaps = 4/76 (5%)
Frame = +3
Query: 124 EVKTKKVDEDHVDDINDNPEVFEHSRGKRRLKQSGSESETVKVTTKKKLLVE----SDEV 179
+VKTKKVD+ V+ +D E + + + + S+ E V+ KKK + S EV
Sbjct: 333 KVKTKKVDDAAVEVEDDKKEKKKKKKKDKENGAAASDEEKVEKEKKKKHKEKGEDGSPEV 512
Query: 180 SDFKPSRAKRRVSSDI 195
+ K + +S++
Sbjct: 513 EKSDKKKKKHKETSEV 560
>TC76961 similar to GP|1841870|gb|AAB68395.1| elongation factor 1-beta
{Pimpinella brachycarpa}, partial (95%)
Length = 815
Score = 28.5 bits (62), Expect = 6.4
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 14/83 (16%)
Frame = +1
Query: 116 ISSENTSVEVKTKKVDEDHV--------------DDINDNPEVFEHSRGKRRLKQSGSES 161
+S E + V V++ V ED V DD +D+ ++F + + K + +
Sbjct: 256 VSGEGSGVTVESSLVAEDAVATPPVSDTKAAEAEDDDDDDVDLFGEETEEEK-KAAEERA 432
Query: 162 ETVKVTTKKKLLVESDEVSDFKP 184
TVK + KKK +S + D KP
Sbjct: 433 ATVKASGKKKESGKSSVLLDVKP 501
>TC91589 similar to GP|5531416|emb|CAB50925.1 translocon Tic40 {Pisum
sativum}, partial (68%)
Length = 1105
Score = 28.1 bits (61), Expect = 8.3
Identities = 21/78 (26%), Positives = 33/78 (41%)
Frame = +1
Query: 117 SSENTSVEVKTKKVDEDHVDDINDNPEVFEHSRGKRRLKQSGSESETVKVTTKKKLLVES 176
S +V++ KV+ DIN EV + K+ S ETV+ ++ +
Sbjct: 163 SQSTVTVDIPATKVESSPGPDINVKEEVEVKNEPKKSAFVDVSPEETVQQNAFER-FKDI 339
Query: 177 DEVSDFKPSRAKRRVSSD 194
D S FK +RA S +
Sbjct: 340 DSSSSFKEARAPAEASQN 393
>BE203403 similar to GP|12322785|gb unknown protein; 24439-25635 {Arabidopsis
thaliana}, partial (47%)
Length = 575
Score = 28.1 bits (61), Expect = 8.3
Identities = 16/35 (45%), Positives = 17/35 (47%)
Frame = +2
Query: 378 WDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYN 412
WD L +YLKE K VVL N H E D N
Sbjct: 146 WDVLLKSYLKEFSKDDSVVLYLLTNPYHTERDFGN 250
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,226,020
Number of Sequences: 36976
Number of extensions: 182639
Number of successful extensions: 713
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 700
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 706
length of query: 507
length of database: 9,014,727
effective HSP length: 100
effective length of query: 407
effective length of database: 5,317,127
effective search space: 2164070689
effective search space used: 2164070689
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0278.2


