

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0276.6
(483 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
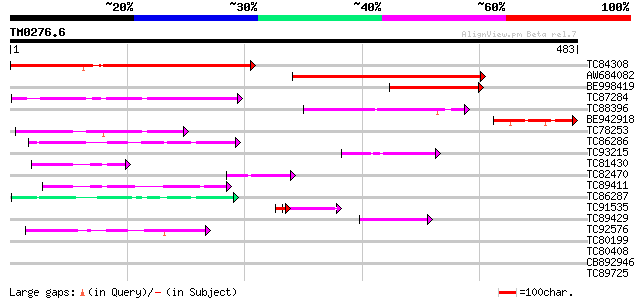
Score E
Sequences producing significant alignments: (bits) Value
TC84308 similar to PIR|T47773|T47773 hypothetical protein F24I3.... 362 e-100
AW684082 homologue to PIR|T47773|T47 hypothetical protein F24I3.... 308 2e-84
BE998419 similar to PIR|T47773|T477 hypothetical protein F24I3.2... 151 6e-37
TC87284 weakly similar to GP|11340603|emb|CAC17054. unnamed prot... 99 3e-21
TC88396 weakly similar to GP|18616497|emb|CAD22847. unnamed prot... 88 8e-18
BE942918 homologue to PIR|H84840|H848 hypothetical protein At2g4... 74 1e-13
TC78253 weakly similar to GP|22654983|gb|AAM98084.1 AT5g45110/K1... 65 6e-11
TC86286 similar to GP|6862923|gb|AAF30312.1| unknown protein {Ar... 50 2e-06
TC93215 similar to GP|10177503|dbj|BAB10897. ankyrin repeat prot... 49 3e-06
TC81430 weakly similar to GP|18491179|gb|AAL69492.1 unknown prot... 47 1e-05
TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical prot... 44 1e-04
TC89411 weakly similar to GP|18491179|gb|AAL69492.1 unknown prot... 44 1e-04
TC86287 similar to GP|23617108|dbj|BAC20790. contains ESTs AU056... 44 1e-04
TC91535 similar to PIR|T06521|T06521 pitrilysin (EC 3.4.24.55) -... 31 5e-04
TC89429 similar to GP|15010672|gb|AAK73995.1 AT3g23280/K14B15_17... 41 9e-04
TC92576 similar to GP|21536509|gb|AAM60841.1 unknown {Arabidopsi... 41 9e-04
TC80199 weakly similar to GP|21593112|gb|AAM65061.1 unknown {Ara... 40 0.002
TC80408 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20 ... 40 0.002
CB892946 GP|6630549|gb| putative RING zinc finger protein {Arabi... 40 0.002
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 31 0.002
>TC84308 similar to PIR|T47773|T47773 hypothetical protein F24I3.210 -
Arabidopsis thaliana, partial (42%)
Length = 755
Score = 362 bits (928), Expect = e-100
Identities = 186/212 (87%), Positives = 192/212 (89%), Gaps = 3/212 (1%)
Frame = +2
Query: 1 MSLEDSLRSLSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPP 60
MSLEDSLRSLSLDYLNLLINGQAFSDV FSVEGRLVHAHRCILAARSLFFRKFFCGPDPP
Sbjct: 89 MSLEDSLRSLSLDYLNLLINGQAFSDVVFSVEGRLVHAHRCILAARSLFFRKFFCGPDPP 268
Query: 61 P---PSGNLDSPGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPN 117
PSGN +P G S R GVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPN
Sbjct: 269 SGLDPSGNRVNPSG----SAR-SGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPN 433
Query: 118 CGERACWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLAS 177
CG+R CWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLAS
Sbjct: 434 CGDRGCWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLAS 613
Query: 178 RKQDMHQLWTTCSHLVAKSGLPPEVLAKHLPI 209
RKQDMHQLWTTCSHLVAKSGLPPE +++ +
Sbjct: 614 RKQDMHQLWTTCSHLVAKSGLPPESTLQNISL 709
Score = 39.7 bits (91), Expect = 0.003
Identities = 20/42 (47%), Positives = 29/42 (68%)
Frame = +1
Query: 182 MHQLWTTCSHLVAKSGLPPEVLAKHLPIDIVAKIEELRLKST 223
M ++ CS + + S + AKHLPIDI+AKIEELR+K++
Sbjct: 637 MDHMFPPCSKIRSSSR---KYFAKHLPIDIIAKIEELRIKTS 753
>AW684082 homologue to PIR|T47773|T47 hypothetical protein F24I3.210 -
Arabidopsis thaliana, partial (35%)
Length = 496
Score = 308 bits (790), Expect = 2e-84
Identities = 158/165 (95%), Positives = 162/165 (97%), Gaps = 1/165 (0%)
Frame = +2
Query: 242 HHD-MGAAADLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREVV 300
HHD + AAADLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREVV
Sbjct: 2 HHDHLTAAADLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREVV 181
Query: 301 KALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDILR 360
KALLELGAADVN+P+GP+GKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDILR
Sbjct: 182 KALLELGAADVNFPAGPTGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDILR 361
Query: 361 TLTSDFLFKGAVPGLTHIEPNKLRLCLELVQSAALVMSREEGNAN 405
TLTSDFLFKGAVPGLTHIEPNKLRLCLELVQSAALVMSREEGN N
Sbjct: 362 TLTSDFLFKGAVPGLTHIEPNKLRLCLELVQSAALVMSREEGNNN 496
>BE998419 similar to PIR|T47773|T477 hypothetical protein F24I3.210 -
Arabidopsis thaliana, partial (17%)
Length = 243
Score = 151 bits (381), Expect = 6e-37
Identities = 74/80 (92%), Positives = 77/80 (95%)
Frame = +2
Query: 324 HIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDILRTLTSDFLFKGAVPGLTHIEPNKL 383
H+AAEMVSP+MVAVLLDHHADP VRTVDGVTPLDILRTLTSDFLFKGAVPGL HIEPNKL
Sbjct: 2 HVAAEMVSPEMVAVLLDHHADPTVRTVDGVTPLDILRTLTSDFLFKGAVPGLNHIEPNKL 181
Query: 384 RLCLELVQSAALVMSREEGN 403
RLCLELVQSAALV+SREE N
Sbjct: 182 RLCLELVQSAALVLSREENN 241
>TC87284 weakly similar to GP|11340603|emb|CAC17054. unnamed protein product
{Zea mays}, partial (16%)
Length = 1027
Score = 99.4 bits (246), Expect = 3e-21
Identities = 68/198 (34%), Positives = 101/198 (50%), Gaps = 1/198 (0%)
Frame = +1
Query: 2 SLEDSLRSLSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFC-GPDPP 60
SLE L + DY D VE V HRCILA+RS FF + F G D
Sbjct: 481 SLEKLLSDVDYDYC----------DAEILVEEIPVGIHRCILASRSQFFHELFKKGKDGE 630
Query: 61 PPSGNLDSPGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGE 120
G G PR ++P SVGYE F++ L +LY+G++ P + C +
Sbjct: 631 VKDGK----GKPRYLMKE---LVPYGSVGYEAFIVFLHYLYTGKLKAPPPEVT---TCVD 780
Query: 121 RACWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQ 180
AC H C A++ AL+ + A+ F +++LAL+ Q+ L + V+KA +EDV+ +L+A+
Sbjct: 781 EACIHDSCRPAINFALELMYASSTFQMKELALVFQRCLLNYVDKALVEDVIPILMAAHHC 960
Query: 181 DMHQLWTTCSHLVAKSGL 198
QL + C VA+S +
Sbjct: 961 KQDQLLSHCIQRVARSDM 1014
>TC88396 weakly similar to GP|18616497|emb|CAD22847. unnamed protein product
{Triticum aestivum}, partial (32%)
Length = 1399
Score = 87.8 bits (216), Expect = 8e-18
Identities = 56/143 (39%), Positives = 84/143 (58%), Gaps = 2/143 (1%)
Frame = +2
Query: 251 LEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREVVKALLELGAAD 310
+ D+ IR++ +ALDS DVEL+KL++ + LD+A AL YA C +VV+ +L LG AD
Sbjct: 131 VSDKSIRKILKALDSDDVELLKLLLDESSVTLDDAYALHYACAYCDSKVVQEVLTLGLAD 310
Query: 311 VNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDILRTLT--SDFLF 368
+ P G T LH+AA P ++ LL + A + T+DG T L I + LT D+
Sbjct: 311 I-LLKNPRGYTVLHVAARRKDPSILVALLKNGACASETTLDGQTALSICQRLTRRKDYHE 487
Query: 369 KGAVPGLTHIEPNKLRLCLELVQ 391
K A T E +K RLC+++++
Sbjct: 488 KTA----TGKESHKDRLCVDVLE 544
>BE942918 homologue to PIR|H84840|H848 hypothetical protein At2g41370
[imported] - Arabidopsis thaliana, partial (4%)
Length = 340
Score = 74.3 bits (181), Expect = 1e-13
Identities = 46/81 (56%), Positives = 55/81 (67%), Gaps = 10/81 (12%)
Frame = +3
Query: 413 NAPCSAATPIYPP--MNEDHNSSSSNANLNLDSRLVYLNLGA-AQM------SGDDDSSH 463
NA S+ T +YP MNEDH+ S +N N+ DSRLVYLNLGA QM SGDDD+
Sbjct: 3 NATGSSGTNMYPHHNMNEDHHHSHNNNNM--DSRLVYLNLGANTQMSTSRMDSGDDDN-- 170
Query: 464 GSHREAMNQAMYHHH-HSHDY 483
+HREA+N +MYHHH H HDY
Sbjct: 171 -THREAINNSMYHHHSHGHDY 230
>TC78253 weakly similar to GP|22654983|gb|AAM98084.1 AT5g45110/K17O22_11
{Arabidopsis thaliana}, partial (32%)
Length = 1669
Score = 65.1 bits (157), Expect = 6e-11
Identities = 45/152 (29%), Positives = 68/152 (44%), Gaps = 5/152 (3%)
Frame = +2
Query: 6 SLRSLSLDYLNLLINGQA-FSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSG 64
SL LS + LLI+ + D VEG V HRCIL +RS FF + F
Sbjct: 1004 SLNKLSSNLEQLLIDSDYDYGDADIIVEGIPVRIHRCILGSRSKFFHEIF---------- 1153
Query: 65 NLDSPGGPRVNSPR----PGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGE 120
G N R ++P VGYE FL+ L ++YSG++ P + C +
Sbjct: 1154 KRSKDKGLSKNEGRLKYCLSDLLPYGKVGYEAFLIFLSYVYSGKLKPSPMEVS---TCVD 1324
Query: 121 RACWHTHCTSAVDLALDTLAAARYFGVEQLAL 152
C H C A++ A++ + A+ F + + +
Sbjct: 1325 NVCAHDACGPAINFAVELMYASSIFQIPEFGI 1420
>TC86286 similar to GP|6862923|gb|AAF30312.1| unknown protein {Arabidopsis
thaliana}, partial (91%)
Length = 1729
Score = 50.1 bits (118), Expect = 2e-06
Identities = 50/180 (27%), Positives = 77/180 (42%)
Frame = +3
Query: 17 LLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNS 76
LL NG+ SDV+F V+G + AH+ +LAARS FR GP + +
Sbjct: 882 LLENGKG-SDVSFEVDGEVFTAHKLVLAARSPVFRAQLFGPMRDQSTQS----------- 1025
Query: 77 PRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLAL 136
I V + VF +L F+Y + P+ E +T + + +A
Sbjct: 1026 ------IKVEDMEAPVFKALLHFMY----------WDSLPDMQELTGMNTKWATTL-MAQ 1154
Query: 137 DTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTCSHLVAKS 196
LAAA + +E+L L+ + AS+ E +I V L + + QL C +A S
Sbjct: 1155 HLLAAADRYALERLRLICE---ASLCEDVAINTVATTLALAEQHHCFQLKAVCLKFIATS 1325
>TC93215 similar to GP|10177503|dbj|BAB10897. ankyrin repeat protein EMB506
{Arabidopsis thaliana}, partial (31%)
Length = 775
Score = 49.3 bits (116), Expect = 3e-06
Identities = 32/85 (37%), Positives = 45/85 (52%)
Frame = +1
Query: 283 DEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHH 342
D A L YAVE +++ VK L++ DVN G TPLH+A + + D+ +LL +
Sbjct: 16 DGATPLHYAVEVGAKQTVKLLIKYNV-DVNVADN-EGWTPLHVAVQSRNRDIAKILLANG 189
Query: 343 ADPNVRTVDGVTPLDILRTLTSDFL 367
AD + DG T LDI DF+
Sbjct: 190 ADRSTENKDGKTALDISICYGKDFM 264
>TC81430 weakly similar to GP|18491179|gb|AAL69492.1 unknown protein
{Arabidopsis thaliana}, partial (49%)
Length = 1491
Score = 47.4 bits (111), Expect = 1e-05
Identities = 31/85 (36%), Positives = 42/85 (48%)
Frame = +2
Query: 19 INGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSPR 78
+N SDVTF VEG+ +AHR L A S FR F GG R R
Sbjct: 695 VNNATLSDVTFLVEGKRFYAHRICLLASSDAFRAMF--------------DGGYREKDAR 832
Query: 79 PGGVIPVNSVGYEVFLLMLQFLYSG 103
I + ++ +EVF LM++F+Y+G
Sbjct: 833 D---IEIPNIRWEVFELMMRFIYTG 898
>TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (8%)
Length = 1064
Score = 44.3 bits (103), Expect = 1e-04
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Frame = -1
Query: 185 LWTTCSHLVAKSGL-PPEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHGHH 243
LWT S L+ S P+++ LP + + ++ + T RR HHHHHHHHH HH
Sbjct: 206 LWTAISSLLLVSCF*YPQMI--FLPTQVFL*VWKMIYQLTCRRRHHHHHHHHHHHHHHHH 33
>TC89411 weakly similar to GP|18491179|gb|AAL69492.1 unknown protein
{Arabidopsis thaliana}, partial (16%)
Length = 839
Score = 43.9 bits (102), Expect = 1e-04
Identities = 44/161 (27%), Positives = 66/161 (40%)
Frame = +2
Query: 29 FSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSPRPGGVIPVNSV 88
F VEG+ HAHR L A S FR F GG R R I + ++
Sbjct: 47 FLVEGKRFHAHRICLLASSDAFRAMF--------------DGGYREKDARD---IEIPNI 175
Query: 89 GYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALDTLAAARYFGVE 148
++VF LM++F+Y+G V + +++A D L AA + +E
Sbjct: 176 RWQVFELMMRFIYTGSVDV-----------------------TLEIAQDLLRAADQYLLE 286
Query: 149 QLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTC 189
L L + +A V S+E+V + S + L TC
Sbjct: 287 GLKRLCEYTIAQHV---SLENVSSMYELSEAFNATSLRHTC 400
>TC86287 similar to GP|23617108|dbj|BAC20790. contains ESTs AU056959(S21019)
AU056960(S21019)~similar to Arabidopsis thaliana
chromosome 5, partial (82%)
Length = 1302
Score = 43.9 bits (102), Expect = 1e-04
Identities = 50/194 (25%), Positives = 79/194 (39%)
Frame = +3
Query: 2 SLEDSLRSLSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPP 61
S+ S ++ + LL +G++ SDV F V G + AH+ +LAARS FR GP
Sbjct: 732 SIPISPSTIGHQFGKLLESGKS-SDVNFEVNGEIFAAHKLVLAARSPVFRAQLFGP---- 896
Query: 62 PSGNLDSPGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGER 121
+ I V + VF +L +Y +P E G
Sbjct: 897 -------------MKDQNTQCIKVEDIEAPVFKALLHVIYWDS---LPDMQE---LTGIN 1019
Query: 122 ACWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQD 181
+ W T +A LAAA + +++L L+ + AS+ E +I V L + +
Sbjct: 1020SKWATTL-----MAQHLLAAADRYALDRLRLMCE---ASLCEDVAINTVATTLALAEQHH 1175
Query: 182 MHQLWTTCSHLVAK 195
QL C +A+
Sbjct: 1176CFQLKAVCLKFIAR 1217
Score = 29.6 bits (65), Expect = 2.7
Identities = 12/33 (36%), Positives = 16/33 (48%)
Frame = +1
Query: 208 PIDIVAKIEELRLKSTLARRSLIPHHHHHHHHH 240
PI ++ + +R I HHH HHHHH
Sbjct: 142 PIHLLVALIWVRFFEKQRGHPQIHHHHLHHHHH 240
Score = 28.9 bits (63), Expect = 4.6
Identities = 9/13 (69%), Positives = 9/13 (69%)
Frame = +1
Query: 231 PHHHHHHHHHGHH 243
P HHHH HH HH
Sbjct: 202 PQIHHHHLHHHHH 240
>TC91535 similar to PIR|T06521|T06521 pitrilysin (EC 3.4.24.55) - garden
pea, partial (26%)
Length = 1146
Score = 31.2 bits (69), Expect(2) = 5e-04
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +2
Query: 227 RSLIPHHHHHHHH 239
RSL +HHHHHHH
Sbjct: 26 RSLYAYHHHHHHH 64
Score = 30.0 bits (66), Expect(2) = 5e-04
Identities = 15/50 (30%), Positives = 21/50 (42%)
Frame = +3
Query: 233 HHHHHHHHGHHDMGAAADLEDQKIRRMRRALDSSDVELVKLMVMGEGLNL 282
HHHHH HH H + + +L S + + +V G GL L
Sbjct: 126 HHHHHRHHRHSPSSISTRFRSNRFFLSSSSLSFSSPQRERRVVYG-GLGL 272
Score = 30.0 bits (66), Expect = 2.1
Identities = 11/19 (57%), Positives = 12/19 (62%)
Frame = +3
Query: 222 STLARRSLIPHHHHHHHHH 240
S+L SL HHHH HH H
Sbjct: 99 SSLTNLSLPHHHHHRHHRH 155
>TC89429 similar to GP|15010672|gb|AAK73995.1 AT3g23280/K14B15_17
{Arabidopsis thaliana}, partial (57%)
Length = 1190
Score = 41.2 bits (95), Expect = 9e-04
Identities = 25/62 (40%), Positives = 34/62 (54%)
Frame = +1
Query: 299 VVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDI 358
V K L+ELGA Y G +PLH AA+ +V +LL H A+P + D TPL++
Sbjct: 340 VAKTLIELGANVNAYRPGRHAGSPLHHAAKRGLESIVKLLLLHGANPLLMNDDCQTPLEV 519
Query: 359 LR 360
R
Sbjct: 520 AR 525
>TC92576 similar to GP|21536509|gb|AAM60841.1 unknown {Arabidopsis
thaliana}, partial (66%)
Length = 905
Score = 41.2 bits (95), Expect = 9e-04
Identities = 43/164 (26%), Positives = 68/164 (41%), Gaps = 6/164 (3%)
Frame = +3
Query: 14 YLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFR-KFFCGPDPPPPSGNLDSPGGP 72
+ L+ SDVTF V G H+ +LAARS FR KFF G D +D+
Sbjct: 351 HFGALLENNEGSDVTFDVAGEKFPGHKLVLAARSPEFRSKFFDGTD-------MDTQ--- 500
Query: 73 RVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTS-- 130
+ V + +VF ML F+Y R + E + TS
Sbjct: 501 ---------EVVVTDLEPKVFKAMLHFIY-------------RDSLTEEVDMVSSTTSSD 614
Query: 131 ---AVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVM 171
+ L L AA +G+E+L L+ + +L + +S+ +++
Sbjct: 615 FPVSETLIAKLLGAADKYGLERLRLMCESRLCKDIGVSSVANIL 746
>TC80199 weakly similar to GP|21593112|gb|AAM65061.1 unknown {Arabidopsis
thaliana}, partial (41%)
Length = 934
Score = 40.4 bits (93), Expect = 0.002
Identities = 12/17 (70%), Positives = 14/17 (81%)
Frame = +3
Query: 227 RSLIPHHHHHHHHHGHH 243
R ++ HHHHHHHHH HH
Sbjct: 339 RQIVLHHHHHHHHHHHH 389
Score = 37.7 bits (86), Expect = 0.010
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +3
Query: 224 LARRSLIPHHHHHHHHHGH 242
L R+ ++ HHHHHHHHH H
Sbjct: 333 LPRQIVLHHHHHHHHHHHH 389
Score = 36.2 bits (82), Expect = 0.029
Identities = 11/13 (84%), Positives = 11/13 (84%)
Frame = +3
Query: 233 HHHHHHHHGHHDM 245
HHHHHHHH HH M
Sbjct: 354 HHHHHHHHHHHHM 392
Score = 32.7 bits (73), Expect = 0.32
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = +3
Query: 218 LRLKSTLARRSLIPHHHHHHHHH 240
L L++ + L HHHHHHHHH
Sbjct: 318 LHLRNLPRQIVLHHHHHHHHHHH 386
Score = 28.9 bits (63), Expect = 4.6
Identities = 11/18 (61%), Positives = 11/18 (61%), Gaps = 4/18 (22%)
Frame = +3
Query: 230 IPHHH----HHHHHHGHH 243
I HHH HHHHHH H
Sbjct: 231 IHHHHQLCIHHHHHHLRH 284
>TC80408 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20
{Arabidopsis thaliana}, partial (22%)
Length = 821
Score = 40.0 bits (92), Expect = 0.002
Identities = 39/165 (23%), Positives = 64/165 (38%), Gaps = 21/165 (12%)
Frame = +3
Query: 236 HHHHHGHHDMGAAADLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEA---------- 285
H+ +H M + + ++DVE K ++ + +++DE
Sbjct: 288 HNSQLRNHTMNHLTISTEDSFASLLELAANNDVEGFKRLIEYDPMSVDEVGLWYGRRKGS 467
Query: 286 --------LALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSP---DM 334
L A S +V+K + L D+N P G T LH AA + D
Sbjct: 468 KQMVNEQRTPLMVAATYGSIDVMKLIFSLSDVDINRPCGLDKSTALHCAASGGAENAVDA 647
Query: 335 VAVLLDHHADPNVRTVDGVTPLDILRTLTSDFLFKGAVPGLTHIE 379
V +LL ADPN +G P+D++ L K ++ L I+
Sbjct: 648 VKLLLAAGADPNSVDANGDRPIDVIVYSPKLELVKNSLEELLQID 782
>CB892946 GP|6630549|gb| putative RING zinc finger protein {Arabidopsis
thaliana}, partial (1%)
Length = 746
Score = 40.0 bits (92), Expect = 0.002
Identities = 12/14 (85%), Positives = 12/14 (85%)
Frame = -2
Query: 232 HHHHHHHHHGHHDM 245
HHHHHHHHH HH M
Sbjct: 229 HHHHHHHHHNHHHM 188
Score = 35.8 bits (81), Expect = 0.038
Identities = 11/14 (78%), Positives = 12/14 (85%)
Frame = -2
Query: 230 IPHHHHHHHHHGHH 243
I HHHHHHHH+ HH
Sbjct: 232 IHHHHHHHHHNHHH 191
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 31.2 bits (69), Expect = 0.93
Identities = 10/17 (58%), Positives = 11/17 (63%)
Frame = -3
Query: 227 RSLIPHHHHHHHHHGHH 243
R + HHHHHH H HH
Sbjct: 121 RHRLHHHHHHHTFHHHH 71
Score = 30.8 bits (68), Expect(2) = 0.002
Identities = 9/14 (64%), Positives = 10/14 (71%)
Frame = -3
Query: 232 HHHHHHHHHGHHDM 245
HHHHHHH HH +
Sbjct: 109 HHHHHHHTFHHHHL 68
Score = 28.5 bits (62), Expect = 6.0
Identities = 10/17 (58%), Positives = 10/17 (58%)
Frame = -3
Query: 226 RRSLIPHHHHHHHHHGH 242
R L HHHHH HH H
Sbjct: 121 RHRLHHHHHHHTFHHHH 71
Score = 28.1 bits (61), Expect(2) = 0.002
Identities = 10/30 (33%), Positives = 17/30 (56%)
Frame = -2
Query: 208 PIDIVAKIEELRLKSTLARRSLIPHHHHHH 237
P ++ I++++ K R + HHHHHH
Sbjct: 299 PTKLLQIIKKIKSKPKKRLRKQLNHHHHHH 210
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,710,960
Number of Sequences: 36976
Number of extensions: 311046
Number of successful extensions: 11069
Number of sequences better than 10.0: 289
Number of HSP's better than 10.0 without gapping: 4134
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7834
length of query: 483
length of database: 9,014,727
effective HSP length: 100
effective length of query: 383
effective length of database: 5,317,127
effective search space: 2036459641
effective search space used: 2036459641
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0276.6


