

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
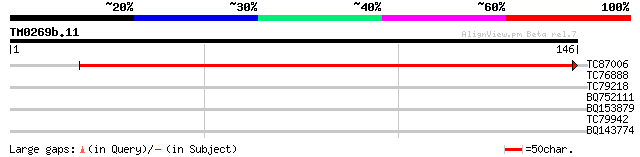
Query= TM0269b.11
(146 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87006 homologue to SP|P06669|CYF_VICFA Apocytochrome F precurs... 239 3e-64
TC76888 similar to PIR|D75542|D75542 hypothetical protein - Dein... 27 2.2
TC79218 similar to SP|P45432|CSN1_ARATH COP9 signalosome complex... 27 2.9
BQ752111 similar to PIR|JQ2282|JQ22 negatively phytochrome regul... 26 5.0
BQ153879 similar to PIR|D86412|D864 protein F1K23.6 [imported] -... 25 8.5
TC79942 similar to GP|8927657|gb|AAF82148.1| EST gb|N38213 comes... 25 8.5
BQ143774 25 8.5
>TC87006 homologue to SP|P06669|CYF_VICFA Apocytochrome F precursor. [Broad
bean] {Vicia faba}, partial (78%)
Length = 1548
Score = 239 bits (610), Expect = 3e-64
Identities = 114/128 (89%), Positives = 123/128 (96%)
Frame = +1
Query: 19 KRGALNVGAVLILPEGFELAPTDRISPEIKEKMGNLSFQSYRPTKKNILVVGPVPGQKYS 78
K+GALNVGAVLILPEGFELAP DRISPE+KEK+GNLSFQSYRPTKKNILVVGPVPG+KYS
Sbjct: 97 KKGALNVGAVLILPEGFELAPPDRISPEVKEKIGNLSFQSYRPTKKNILVVGPVPGKKYS 276
Query: 79 EITFPILSPDPATNRDVNFLKYPIYVGGNRGRGQIYPDGSKSNNNVYNATKSGIINKIIR 138
EITFPILSPDPAT RDV+FLKYPIYVGGNRGRGQIYPDGSKSNNNVYNAT +GI+NKIIR
Sbjct: 277 EITFPILSPDPATKRDVHFLKYPIYVGGNRGRGQIYPDGSKSNNNVYNATATGIVNKIIR 456
Query: 139 KDKGGIHL 146
K+KGG +
Sbjct: 457 KEKGGYEI 480
>TC76888 similar to PIR|D75542|D75542 hypothetical protein - Deinococcus
radiodurans (strain R1), partial (57%)
Length = 1749
Score = 27.3 bits (59), Expect = 2.2
Identities = 13/16 (81%), Positives = 13/16 (81%)
Frame = +2
Query: 1 AVVRKVDLFGNLRILA 16
A VRKVDLFGN RI A
Sbjct: 1622 AAVRKVDLFGNPRIYA 1669
>TC79218 similar to SP|P45432|CSN1_ARATH COP9 signalosome complex subunit 1
(CSN complex subunit 1) (COP11 protein) (FUSCA protein
FUS6)., partial (90%)
Length = 1746
Score = 26.9 bits (58), Expect = 2.9
Identities = 18/58 (31%), Positives = 26/58 (44%), Gaps = 6/58 (10%)
Frame = +1
Query: 40 TDRISPEIKEKM------GNLSFQSYRPTKKNILVVGPVPGQKYSEITFPILSPDPAT 91
TD + P I K+ NL + Y+ + L GP G Y+E+ I + D AT
Sbjct: 712 TDALDPIITSKLRCAAGLANLEAKKYKLAARKFLEAGPELGSTYNEV---IAAQDVAT 876
>BQ752111 similar to PIR|JQ2282|JQ22 negatively phytochrome regulated protein
I - swollen duckweed, partial (17%)
Length = 737
Score = 26.2 bits (56), Expect = 5.0
Identities = 12/39 (30%), Positives = 21/39 (53%)
Frame = +1
Query: 14 ILANGKRGALNVGAVLILPEGFELAPTDRISPEIKEKMG 52
+L + GAL V ++++ P + AP D IS + + G
Sbjct: 457 LLGGVEAGALRVRSLILTPAALQWAPVDIISVHLADAHG 573
>BQ153879 similar to PIR|D86412|D864 protein F1K23.6 [imported] - Arabidopsis
thaliana, partial (6%)
Length = 487
Score = 25.4 bits (54), Expect = 8.5
Identities = 14/56 (25%), Positives = 27/56 (48%)
Frame = -3
Query: 45 PEIKEKMGNLSFQSYRPTKKNILVVGPVPGQKYSEITFPILSPDPATNRDVNFLKY 100
P I L F R +KKNI ++ + +++ +L P+P N+ ++ +Y
Sbjct: 464 P*I*HDCSGLQFGKSRQSKKNIYMLVRIREREFLPKLHSLLLPNPMKNKSISAGQY 297
>TC79942 similar to GP|8927657|gb|AAF82148.1| EST gb|N38213 comes from this
gene. {Arabidopsis thaliana}, partial (9%)
Length = 879
Score = 25.4 bits (54), Expect = 8.5
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 6/35 (17%)
Frame = +1
Query: 39 PTDRISPEIKEKMGNLSFQ------SYRPTKKNIL 67
P ISP +KEK+G+ Q +YRP K + L
Sbjct: 715 PKSNISPCLKEKLGSCEKQIINCINTYRPNKMSSL 819
>BQ143774
Length = 743
Score = 25.4 bits (54), Expect = 8.5
Identities = 12/27 (44%), Positives = 16/27 (58%)
Frame = +1
Query: 30 ILPEGFELAPTDRISPEIKEKMGNLSF 56
+L G +L T +SPE +EKM N F
Sbjct: 661 LLKFGIKLPGTGWVSPETQEKMANRPF 741
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.141 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,682,271
Number of Sequences: 36976
Number of extensions: 46100
Number of successful extensions: 221
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 219
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 221
length of query: 146
length of database: 9,014,727
effective HSP length: 87
effective length of query: 59
effective length of database: 5,797,815
effective search space: 342071085
effective search space used: 342071085
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0269b.11


