

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0269a.2
(447 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
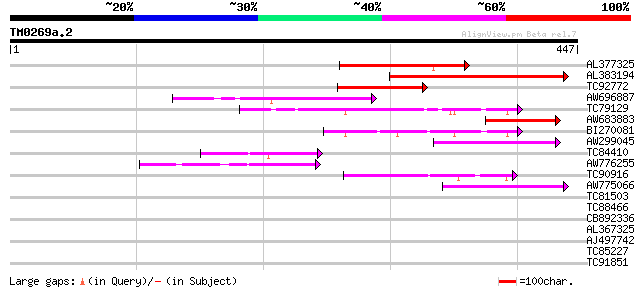
Score E
Sequences producing significant alignments: (bits) Value
AL377325 homologue to GP|3608181|dbj cyclin B {Pisum sativum}, p... 174 8e-44
AL383194 similar to GP|6031209|gb| cyclin {Pisum sativum}, parti... 129 2e-30
TC92772 similar to GP|19347837|gb|AAL86330.1 putative prolyl end... 117 9e-27
AW696887 similar to PIR|T07669|T07 cyclin a1-type mitosis-speci... 94 8e-20
TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.... 74 1e-13
AW683883 similar to EGAD|126998|135 cyclin {Lupinus luteus}, par... 65 5e-11
BI270081 similar to GP|6448480|emb cyclin D1 {Antirrhinum majus}... 55 4e-08
AW299045 weakly similar to GP|18175785|gb putative cyclin {Arabi... 55 5e-08
TC84410 similar to GP|15419009|gb|AAK81695.1 cyclin A2 {Medicago... 51 8e-07
AW776255 weakly similar to PIR|T07672|T07 cyclin a2-type mitosi... 50 2e-06
TC90916 similar to PIR|T09961|T09961 cyclin D-like protein - red... 44 1e-04
AW775066 similar to PIR|T07669|T07 cyclin a1-type mitosis-speci... 42 4e-04
TC81503 similar to GP|6448480|emb|CAB61221.1 cyclin D1 {Antirrhi... 41 0.001
TC88466 similar to GP|6434199|emb|CAB60837.1 CycD3;2 {Lycopersic... 37 0.015
CB892336 homologue to PIR|S56679|S566 mitosis-specific cyclin Cy... 34 0.10
AL367325 homologue to GP|1835260|emb| mitotic cyclin {Sesbania r... 33 0.17
AJ497742 SP|P30278|CG2B G2/mitotic-specific cyclin 2 (B-like cyc... 28 0.68
TC85227 similar to GP|15983773|gb|AAL10483.1 AT4g29060/F19B15_90... 30 1.9
TC91851 weakly similar to PIR|F96673|F96673 hypothetical protein... 30 1.9
>AL377325 homologue to GP|3608181|dbj cyclin B {Pisum sativum}, partial (45%)
Length = 329
Score = 174 bits (440), Expect = 8e-44
Identities = 89/108 (82%), Positives = 96/108 (88%), Gaps = 6/108 (5%)
Frame = +2
Query: 261 RRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVP 320
RRELQLVGIS+MLMA+KYEEIWPPEVNDFVCLSDRAYSHEQIL+MEK ILG+LEWTLTVP
Sbjct: 5 RRELQLVGISAMLMASKYEEIWPPEVNDFVCLSDRAYSHEQILIMEKTILGKLEWTLTVP 184
Query: 321 TPFVFLTRFIKA----SVP-DEG-VTNMAHFLSELGMMHYDTLMYCPS 362
TPFVFL RFIKA +VP D+G + MAHFLSELGMMHY TL YCPS
Sbjct: 185 TPFVFLVRFIKAASVSAVPSDQGDLEMMAHFLSELGMMHYATLRYCPS 328
>AL383194 similar to GP|6031209|gb| cyclin {Pisum sativum}, partial (55%)
Length = 435
Score = 129 bits (324), Expect = 2e-30
Identities = 62/141 (43%), Positives = 89/141 (62%)
Frame = +2
Query: 300 EQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMY 359
+Q+L MEK+IL +L++ L PTP+VFL RF+KA+ D + +MA FL +L ++ Y+ L +
Sbjct: 5 DQMLGMEKLILRKLKFRLNAPTPYVFLIRFLKAAQSDMKLEHMAFFLIDLCLVEYEALAF 184
Query: 360 CPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVV 419
PS++ AS +Y ARCTL +P+W L+ H Y Q+ DCA +++ FH G GKL V
Sbjct: 185 KPSLLCASXLYVARCTLXITPSWTPLLQKHARYEVSQIXDCADMILKFHKAAGKGKLTVA 364
Query: 420 FRKYSDPERGAVAVLPPAKNL 440
+ KYS E G VA P L
Sbjct: 365 YEKYSRKELGGVAAXKPLDRL 427
>TC92772 similar to GP|19347837|gb|AAL86330.1 putative prolyl endopeptidase
{Arabidopsis thaliana}, partial (14%)
Length = 531
Score = 117 bits (293), Expect = 9e-27
Identities = 55/71 (77%), Positives = 62/71 (86%)
Frame = +3
Query: 259 VPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLT 318
VPR+ELQLVGISSML+A+KYEEIW PEVNDFVC+SD AY EQ+LVMEK IL LEW LT
Sbjct: 318 VPRKELQLVGISSMLIASKYEEIWAPEVNDFVCISDNAYVREQVLVMEKTILRNLEWYLT 497
Query: 319 VPTPFVFLTRF 329
VPTP+VFL R+
Sbjct: 498 VPTPYVFLVRY 530
>AW696887 similar to PIR|T07669|T07 cyclin a1-type mitosis-specific -
soybean, partial (33%)
Length = 648
Score = 94.4 bits (233), Expect = 8e-20
Identities = 58/165 (35%), Positives = 97/165 (58%), Gaps = 4/165 (2%)
Frame = +3
Query: 129 PDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNELAAV 188
PD + + +Q +K + K + T++ + S NKP ++ DN+
Sbjct: 174 PDFDYQPTQKLQCRKNPNLKNSATTIPSLDSNL-----NKPVHTKINAKR---DNQHIIQ 329
Query: 189 EYIEDIYKFYKMVENES--RPHC-YMAS-QPEINEKMRAILVDWLIDVHTKFELSLETLY 244
Y+ DI + + +E + RP Y+ + Q I+ MR LVDWL++V +++L ETL+
Sbjct: 330 PYVSDISDYLRTMEMQKKRRPMVGYLENVQRGISSNMRGTLVDWLVEVADEYKLLPETLH 509
Query: 245 LTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDF 289
L+++ +DRFL+++ V R +LQL+G+SSML+A+KYEEI PP+ DF
Sbjct: 510 LSVSYIDRFLSIEPVSRSKLQLLGVSSMLIASKYEEITPPKAVDF 644
>TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.7
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1328
Score = 73.6 bits (179), Expect = 1e-13
Identities = 67/239 (28%), Positives = 112/239 (46%), Gaps = 16/239 (6%)
Frame = +2
Query: 182 DNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLE 241
+ E + +IE +KF + SR + + E + + AI W++ VH +
Sbjct: 257 EEEESIAVFIEHEFKFVPGFDYVSR---FQSRSLESSTREEAIA--WILKVHEYYGFQPL 421
Query: 242 TLYLTINIVDRFLAVKTVPRRE---LQLVGISSMLMAAKYEEIWPPEVNDFVCLSDR-AY 297
T YL++N +DRFL + +P LQL+ ++ + +AAK EE P + DF + +
Sbjct: 422 TAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIF 601
Query: 298 SHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFL----SEL---G 350
IL ME ++L L+W L TP FL+ F + G HF+ +E+
Sbjct: 602 QPRTILRMELLVLTILDWRLRSITPLSFLS-FFACKLDSTG--TFTHFIISRATEIILSN 772
Query: 351 MMHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTD-----YSEEQLMDCARLL 404
+ L Y PS IAA+A+ +A N+ P W+ H + S+E+++ C L+
Sbjct: 773 IQDASFLTYRPSCIAAAAILSA---ANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELI 940
>AW683883 similar to EGAD|126998|135 cyclin {Lupinus luteus}, partial (12%)
Length = 217
Score = 65.1 bits (157), Expect = 5e-11
Identities = 27/59 (45%), Positives = 41/59 (68%)
Frame = +2
Query: 376 LNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVL 434
L +SP W +TLK +T YSEEQL DCA+L+ SFH +L+ +++K+ +R AVA++
Sbjct: 8 LERSPFWTDTLKHYTGYSEEQLRDCAKLMASFHSAAPESRLRAIYKKFCSSDRCAVALI 184
>BI270081 similar to GP|6448480|emb cyclin D1 {Antirrhinum majus}, partial
(50%)
Length = 663
Score = 55.5 bits (132), Expect = 4e-08
Identities = 53/174 (30%), Positives = 82/174 (46%), Gaps = 17/174 (9%)
Frame = +2
Query: 248 NIVDRFLAVKTVPRRE---LQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILV 304
N +DRFL + +P+ LQL+ ++ + +AAK EE P + D + Y E I +
Sbjct: 2 NYMDRFLNSRRLPQTNGWPLQLLSVACLSLAAKMEETLVPSLLDLQ-VEGVKYMFEPITI 178
Query: 305 --MEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSEL------GMMHYDT 356
ME ++L L+W L TPF FL+ F A D T +S +
Sbjct: 179 RRMELLVLSVLDWRLRSVTPFSFLSFF--ACKLDSTSTFTGFLISRATQIILSKIQEASI 352
Query: 357 LMYCPSMIAASAV-YAARCTLNKSPAWNETLKLHTD-----YSEEQLMDCARLL 404
L Y PS IAA+A+ YAA N+ P W+ H + +E+++ C +L+
Sbjct: 353 LAYWPSCIAAAAILYAA----NEIPNWSLVEPEHAESWCEGLRKEKIIGCYQLM 502
>AW299045 weakly similar to GP|18175785|gb putative cyclin {Arabidopsis
thaliana}, partial (24%)
Length = 646
Score = 55.1 bits (131), Expect = 5e-08
Identities = 36/101 (35%), Positives = 51/101 (49%), Gaps = 1/101 (0%)
Frame = -1
Query: 335 PDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKS-PAWNETLKLHTDYS 393
P + +A+FL+EL +M Y L + PSMIAASAV+ AR TL++S WN TL+ + Y
Sbjct: 637 PSVELEYLANFLAELTLMSYGFLNFFPSMIAASAVFLARWTLDQSRHPWNPTLEHYASYK 458
Query: 394 EEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVL 434
L L + + + KY + VAVL
Sbjct: 457 ASDLKATVLALQNLQLNSDDCPYPAIRTKYRQSKFHGVAVL 335
>TC84410 similar to GP|15419009|gb|AAK81695.1 cyclin A2 {Medicago sativa},
partial (23%)
Length = 771
Score = 51.2 bits (121), Expect = 8e-07
Identities = 35/99 (35%), Positives = 49/99 (49%), Gaps = 3/99 (3%)
Frame = +1
Query: 151 LTSVLTARSKAACGLTNKPKE-EIVDIDASDVDNELAAVEYIEDIYKFYKMVE--NESRP 207
L S A+ + L N PK+ +I +IDA D +L + Y DIY ++ E P
Sbjct: 478 LISQAKAKKDSDSKLMNAPKDPDIPNIDADIEDPQLCSF-YAADIYDNLRVAELSXXXHP 654
Query: 208 HCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLT 246
+ Q +I MRAIL+DWL++V F L YLT
Sbjct: 655 NFMETVQRDITXXMRAILIDWLVEVSXXFNLQANXXYLT 771
>AW776255 weakly similar to PIR|T07672|T07 cyclin a2-type mitosis-specific -
soybean, partial (22%)
Length = 704
Score = 49.7 bits (117), Expect = 2e-06
Identities = 45/146 (30%), Positives = 71/146 (47%), Gaps = 3/146 (2%)
Frame = +1
Query: 103 KREAPKPVTKRVTGKPKPVVEVIEISPDEQIKKEKSVQKKKED-SKKKTLTSVLTARSKA 161
K +A K + K + E +++ P E S+ ++ + T +++ + A
Sbjct: 298 KEDAKKELAKDSSTSTMTKKESLQVQPSV----ENSLLSMQDTLNSPNTEINLICEKLSA 465
Query: 162 ACGLTNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHC-YMAS-QPEINE 219
+ GL IVDID+ D+ + Y DIY + E E RP YM + Q +I
Sbjct: 466 SVGLG------IVDIDSKLRDSPIWT-SYAPDIYTNIHVRECERRPLANYMETLQQDITP 624
Query: 220 KMRAILVDWLIDVHTKFELSLETLYL 245
MR ILVDWL++V +F+L +TLYL
Sbjct: 625 GMRGILVDWLVEVADEFKLVSDTLYL 702
>TC90916 similar to PIR|T09961|T09961 cyclin D-like protein - red goosefoot,
partial (31%)
Length = 886
Score = 44.3 bits (103), Expect = 1e-04
Identities = 38/147 (25%), Positives = 74/147 (49%), Gaps = 10/147 (6%)
Frame = +3
Query: 264 LQLVGISSMLMAAKYEEIWPPEVNDF-VCLSDRAYSHEQILVMEKIILGRLEWTLTVPTP 322
+QL+ ++ + +AAK EE P+ D + S + + I ME ++L L+W + TP
Sbjct: 36 MQLLAVACLSLAAKVEETAVPQPLDLQIGESKFVFEAKTIQRMELLVLSTLKWRMQAITP 215
Query: 323 FVFLTRFIKASVPDEGVTNMAHFLSELGMM------HYDTLMYCPSMIAASAVYAARCTL 376
F F+ F+ + + D+ ++++ +S + D L + PS IAA+ A C +
Sbjct: 216 FSFIECFL-SKIKDDDKSSLSSSISRSTQLISSTIKGLDFLEFKPSEIAAA---VATCVV 383
Query: 377 NKSPAWNETLKLHT--DYSEE-QLMDC 400
++ A + + + T Y E+ +L+ C
Sbjct: 384 GETQAIDSSKSISTLIQYVEKGRLLKC 464
>AW775066 similar to PIR|T07669|T07 cyclin a1-type mitosis-specific -
soybean, partial (31%)
Length = 696
Score = 42.4 bits (98), Expect = 4e-04
Identities = 26/100 (26%), Positives = 47/100 (47%), Gaps = 1/100 (1%)
Frame = +2
Query: 342 MAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKS-PAWNETLKLHTDYSEEQLMDC 400
+ ++L+EL ++ Y+ + + PSM+AAS ++ AR + +L Y +L +C
Sbjct: 20 LCNYLAELSLIDYECIRFLPSMVAASVIFLARFIICPGVHPLTSSLSECLFYKSAELEEC 199
Query: 401 ARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
+L + LK V KY + VA LP + +
Sbjct: 200 VLILHDLYLVRRAASLKAVREKYKQHKFKNVANLPSSPEI 319
>TC81503 similar to GP|6448480|emb|CAB61221.1 cyclin D1 {Antirrhinum majus},
partial (36%)
Length = 1000
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/66 (31%), Positives = 36/66 (53%), Gaps = 3/66 (4%)
Frame = +3
Query: 226 VDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRE---LQLVGISSMLMAAKYEEIW 282
+ W++ V + T YL +N +DRFL + +P+ LQL+ ++ + +AAK EE
Sbjct: 396 IRWILKVQGYYGFQPVTAYLAVNYMDRFLNSRRLPQTNGWPLQLLSVACLSLAAKMEETL 575
Query: 283 PPEVND 288
P + D
Sbjct: 576 VPSLLD 593
>TC88466 similar to GP|6434199|emb|CAB60837.1 CycD3;2 {Lycopersicon
esculentum}, partial (39%)
Length = 692
Score = 37.0 bits (84), Expect = 0.015
Identities = 25/94 (26%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Frame = +2
Query: 202 ENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPR 261
E ++ + ++ + P + E R ++W++ V+ + S T L +N +DRFL
Sbjct: 140 EQQNPLYIFLQTNPVL-ETARRESIEWILKVNAHYSFSALTSVLAVNYLDRFLFSFRFQN 316
Query: 262 RE---LQLVGISSMLMAAKYEEIWPPEVNDF-VC 291
+ QL ++ + +AAK EE P + D VC
Sbjct: 317 EKPWMTQLAAVACLSLAAKMEETHVPLLLDLQVC 418
>CB892336 homologue to PIR|S56679|S566 mitosis-specific cyclin CycIII -
alfalfa, partial (10%)
Length = 860
Score = 34.3 bits (77), Expect = 0.10
Identities = 17/47 (36%), Positives = 23/47 (48%)
Frame = -3
Query: 399 DCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLMPQEA 445
+C+ L+V FH G GKL RKY + A PA L+ E+
Sbjct: 648 ECSSLMVDFHKKAGTGKLTGAHRKYGTSKFSYTAKCEPASFLLENES 508
>AL367325 homologue to GP|1835260|emb| mitotic cyclin {Sesbania rostrata},
partial (4%)
Length = 126
Score = 33.5 bits (75), Expect = 0.17
Identities = 16/22 (72%), Positives = 18/22 (81%), Gaps = 2/22 (9%)
Frame = +1
Query: 1 MASRPVVPQ--QPRGDAVVGGG 20
MASRP+V Q QPRGDA +GGG
Sbjct: 61 MASRPIVQQHQQPRGDAALGGG 126
>AJ497742 SP|P30278|CG2B G2/mitotic-specific cyclin 2 (B-like cyclin)
(CycMs2) (Fragment). [Alfalfa] {Medicago sativa},
partial (18%)
Length = 532
Score = 28.1 bits (61), Expect(2) = 0.68
Identities = 15/35 (42%), Positives = 17/35 (47%)
Frame = +1
Query: 403 LLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPA 437
L+V FH G GKL V RKY + A PA
Sbjct: 49 LMVGFHQKAGAGKLTGVHRKYGSAKFSFTAKCEPA 153
Score = 21.9 bits (45), Expect(2) = 0.68
Identities = 7/11 (63%), Positives = 11/11 (99%)
Frame = +2
Query: 389 HTDYSEEQLMD 399
HT+YSE+QL++
Sbjct: 8 HTNYSEDQLLE 40
>TC85227 similar to GP|15983773|gb|AAL10483.1 AT4g29060/F19B15_90 {Arabidopsis
thaliana}, partial (63%)
Length = 3731
Score = 30.0 bits (66), Expect = 1.9
Identities = 21/86 (24%), Positives = 38/86 (43%)
Frame = +3
Query: 104 REAPKPVTKRVTGKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAAC 163
+E + +V P+ V E P+E +KKE ++ +KED K RS+
Sbjct: 2304 KELVDDIAMQVAACPQVEYVVTEDVPEEFLKKETEIEMQKEDLASKP----EQIRSRIVE 2471
Query: 164 GLTNKPKEEIVDIDASDVDNELAAVE 189
G K E++ ++ + N+ V+
Sbjct: 2472 GRIRKRLEDLALLEQPYIKNDKVTVK 2549
>TC91851 weakly similar to PIR|F96673|F96673 hypothetical protein F13O11.30
[imported] - Arabidopsis thaliana, partial (12%)
Length = 981
Score = 30.0 bits (66), Expect = 1.9
Identities = 54/222 (24%), Positives = 88/222 (39%), Gaps = 8/222 (3%)
Frame = +3
Query: 73 AVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVE---VIEISP 129
A A + ++ P+ AP A P V+ + KPVTK + P P+ +E SP
Sbjct: 99 ASMASKSSTRSKPSDNSNKAPPATPKVS----KVSKPVTKSASESPSPLQNSRLSVEKSP 266
Query: 130 DEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACG-----LTNKPKEEIVDIDASDVDNE 184
+ + +V++K S K T T +AA G N +E++ + E
Sbjct: 267 -RSVNSKPAVERK---SAKATATPPDKQAPRAAKGSELQNQLNVAQEDLKKAKEQILQAE 434
Query: 185 LAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLY 244
V+ I+++ + ++ E E NEK++ LV K E +E
Sbjct: 435 KEKVKAIDELKEAQRVAE-------------EANEKLQEALV---AQKQAKEESEIE--- 557
Query: 245 LTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEV 286
+F AV EL+ GI ++ K EE W E+
Sbjct: 558 -------KFRAV------ELEQAGIETV---NKKEEEWQKEL 635
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,662,597
Number of Sequences: 36976
Number of extensions: 170936
Number of successful extensions: 1075
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 993
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1060
length of query: 447
length of database: 9,014,727
effective HSP length: 99
effective length of query: 348
effective length of database: 5,354,103
effective search space: 1863227844
effective search space used: 1863227844
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0269a.2


