

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0266.20
(284 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
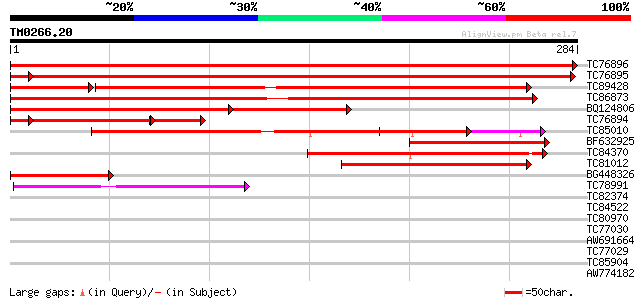
Score E
Sequences producing significant alignments: (bits) Value
TC76896 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 536 e-153
TC76895 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 501 e-144
TC89428 homologue to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus l... 231 3e-74
TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 262 1e-70
BQ124806 similar to SP|P42762|ERD1 ERD1 protein chloroplast pre... 183 4e-60
TC76894 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 127 1e-41
TC85010 weakly similar to PIR|T40514|T40514 Chaperonin hsp78p - ... 146 8e-36
BF632925 similar to GP|15425870|gb| heat shock protein 78 {Candi... 80 9e-16
TC84370 weakly similar to PIR|T39572|T39572 probable proteinase ... 80 9e-16
TC81012 similar to PIR|T51523|T51523 clpB heat shock protein-lik... 78 3e-15
BG448326 similar to PIR|T51523|T51 clpB heat shock protein-like ... 75 2e-14
TC78991 similar to GP|9759268|dbj|BAB09589.1 101 kDa heat shock ... 57 8e-09
TC82374 similar to GP|21689765|gb|AAM67526.1 putative arsA-like ... 32 0.28
TC84522 similar to GP|13161354|dbj|BAB32945. putative calcium se... 32 0.36
TC80970 similar to SP|O24357|G6PC_SPIOL Glucose-6-phosphate 1-de... 31 0.62
TC77030 similar to GP|7406669|emb|CAB85628.1 putative ripening-r... 30 1.1
AW691664 similar to PIR|T52061|T52 tRNA isopentenyltransferase (... 29 1.8
TC77029 similar to GP|7406669|emb|CAB85628.1 putative ripening-r... 29 1.8
TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [i... 29 2.3
AW774182 homologue to PIR|F84674|F84 probable AAA-type ATPase [i... 29 2.3
>TC76896 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], partial (35%)
Length = 1348
Score = 536 bits (1382), Expect = e-153
Identities = 272/284 (95%), Positives = 282/284 (98%)
Frame = +1
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG
Sbjct: 151 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 330
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN
Sbjct: 331 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 510
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
VGSSVIEKGGR+IGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK
Sbjct: 511 VGSSVIEKGGRRIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 690
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
LEVKEIADIMLKEVF+RLKTKEI+LSVTERFR+RVV+EGY+PSYGARPLRRAIMRLLEDS
Sbjct: 691 LEVKEIADIMLKEVFERLKTKEIELSVTERFRERVVDEGYNPSYGARPLRRAIMRLLEDS 870
Query: 241 MAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSGAPESLPEALP 284
MAEKMLA EIKEGDSVI+DADSDG VIVLNGS+GAP+SLP+ALP
Sbjct: 871 MAEKMLAREIKEGDSVIVDADSDGNVIVLNGSTGAPDSLPDALP 1002
>TC76895 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], complete
Length = 3425
Score = 501 bits (1291), Expect(2) = e-144
Identities = 255/273 (93%), Positives = 264/273 (96%)
Frame = +1
Query: 11 GKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 70
GKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR
Sbjct: 2173 GKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 2352
Query: 71 PYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG 130
PYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG
Sbjct: 2353 PYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG 2532
Query: 131 RKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIM 190
R+IGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIM
Sbjct: 2533 RRIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIM 2712
Query: 191 LKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEI 250
L+EVF RLK KEI+L VTERFRDRVV+EGY+PSYGARPLRRAIMRLLEDSMAEKMLA EI
Sbjct: 2713 LREVFQRLKNKEIELQVTERFRDRVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREI 2892
Query: 251 KEGDSVIIDADSDGKVIVLNGSSGAPESLPEAL 283
KEGDSVI+D DS+G VIVLNG+SG ESLPEAL
Sbjct: 2893 KEGDSVIVDVDSEGNVIVLNGTSGTQESLPEAL 2991
Score = 27.3 bits (59), Expect(2) = e-144
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = +3
Query: 1 SFIFSGPTGVGK 12
SFIFSGPTGVG+
Sbjct: 2142 SFIFSGPTGVGE 2177
>TC89428 homologue to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus lunatus},
partial (32%)
Length = 1227
Score = 231 bits (590), Expect(2) = 3e-74
Identities = 117/218 (53%), Positives = 160/218 (72%)
Frame = +2
Query: 44 VSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDS 103
VS+L+G+PPGYVGY EGGQLTE VRRRPY+VVLFDEIEKAH DVFN++LQ+L+DGR+TDS
Sbjct: 317 VSRLVGAPPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHHDVFNILLQLLDDGRITDS 496
Query: 104 KGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRP 163
+GRTV F N +LIMTSN+GS I + D+K + Y+++K V E +Q FRP
Sbjct: 497 QGRTVSFTNCVLIMTSNIGSHHILE-----TLSSTQDDKIAVYDQMKRQVVELARQTFRP 661
Query: 164 EFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPS 223
EF+NR+DE IVF+ L E+ +I ++ ++ V RLK K+IDL TE + G+DP+
Sbjct: 662 EFMNRIDEYIVFQPLDSSEISKIVELQMERVKGRLKQKKIDLHYTEEAVKLLGVLGFDPN 841
Query: 224 YGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 261
+GARP++R I +L+E+ +A +L G+ KE DS+I+DAD
Sbjct: 842 FGARPVKRVIQQLVENEIAMGVLRGDFKEEDSIIVDAD 955
Score = 64.7 bits (156), Expect(2) = 3e-74
Identities = 27/42 (64%), Positives = 36/42 (85%)
Frame = +1
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERH 42
SF+F GPTGVGK+EL K LA+Y F +E A++R+DMSE+ME+H
Sbjct: 187 SFMFMGPTGVGKTELGKALANYLFNTENALVRIDMSEYMEKH 312
>TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (53%)
Length = 1475
Score = 262 bits (670), Expect = 1e-70
Identities = 130/264 (49%), Positives = 186/264 (70%)
Frame = +2
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SF+F GPTGVGK+ELAK LA F E ++R+DMSE+ME+H+VS+LIG+PPGYVG+ EG
Sbjct: 704 SFLFLGPTGVGKTELAKALAEQLFDDENQLVRIDMSEYMEQHSVSRLIGAPPGYVGHEEG 883
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPY+VVLFDE+EKAH VFN +LQ+L+DGRLTD +GRTVDF+NT++IMTSN
Sbjct: 884 GQLTEAVRRRPYSVVLFDEVEKAHTSVFNTLLQVLDDGRLTDGQGRTVDFRNTVIIMTSN 1063
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
+G+ + G + + V +E++++FRPE LNRLDE++VF L+
Sbjct: 1064LGAEHLLSG----------LSGKCTMQAARDRVMQEVRRHFRPELLNRLDEVVVFDPLSH 1213
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
+++++A + +K+V RL + I L+VT+ D ++ E YDP YGARP+R+ + + +
Sbjct: 1214EQLRKVARLQMKDVASRLAERGIALAVTDAALDYILAESYDPVYGARPIRKWLEKKVVTE 1393
Query: 241 MAEKMLAGEIKEGDSVIIDADSDG 264
++ ++ EI E + IDA G
Sbjct: 1394LSRMLIREEIDENTTGYIDAGPKG 1465
>BQ124806 similar to SP|P42762|ERD1 ERD1 protein chloroplast precursor.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (20%)
Length = 679
Score = 183 bits (464), Expect(2) = 4e-60
Identities = 85/112 (75%), Positives = 102/112 (90%)
Frame = +2
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
+ +F GPTGVGK+ELAK+LA+ YFGSE MIRLDMSE+MERH+VSKL+GSPPGYVGY EG
Sbjct: 74 TLLFCGPTGVGKTELAKSLAACYFGSETNMIRLDMSEYMERHSVSKLLGSPPGYVGYGEG 253
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKN 112
G LTEA+RR+P+TVVLFDEIEKAHPD+FN++LQ++EDG LTDS+GR V FKN
Sbjct: 254 GILTEAIRRKPFTVVLFDEIEKAHPDIFNILLQLMEDGHLTDSQGRRVSFKN 409
Score = 65.9 bits (159), Expect(2) = 4e-60
Identities = 34/61 (55%), Positives = 43/61 (69%)
Frame = +3
Query: 111 KNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLD 170
K L++MTSNVGSS I KG L D+K +SY+ +KS+V EEL+ YFRPE LNR+D
Sbjct: 405 KMHLVVMTSNVGSSAIAKGRHNSMGFLISDDKPTSYSGLKSMVIEELRTYFRPELLNRID 584
Query: 171 E 171
E
Sbjct: 585 E 587
>TC76894 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], partial (71%)
Length = 2413
Score = 127 bits (319), Expect(3) = 1e-41
Identities = 62/63 (98%), Positives = 62/63 (98%)
Frame = +1
Query: 11 GKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 70
GKSELAK LASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR
Sbjct: 2149 GKSELAKALASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 2328
Query: 71 PYT 73
PYT
Sbjct: 2329 PYT 2337
Score = 53.1 bits (126), Expect(3) = 1e-41
Identities = 25/28 (89%), Positives = 25/28 (89%)
Frame = +2
Query: 71 PYTVVLFDEIEKAHPDVFNMMLQILEDG 98
P VVLFDEI KAHPDVFNMMLQILEDG
Sbjct: 2330 PTRVVLFDEIXKAHPDVFNMMLQILEDG 2413
Score = 27.3 bits (59), Expect(3) = 1e-41
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = +3
Query: 1 SFIFSGPTGVGK 12
SFIFSGPTGVG+
Sbjct: 2118 SFIFSGPTGVGE 2153
>TC85010 weakly similar to PIR|T40514|T40514 Chaperonin hsp78p - fission
yeast (Schizosaccharomyces pombe), partial (23%)
Length = 904
Score = 146 bits (369), Expect = 8e-36
Identities = 79/203 (38%), Positives = 124/203 (60%), Gaps = 13/203 (6%)
Frame = +3
Query: 42 HTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLT 101
HT+S+LIG+P GYVGY + GQLTEAVRR+PY V+LFDE EKAH D+ ++LQ+L++G LT
Sbjct: 9 HTISRLIGAPSGYVGYEDAGQLTEAVRRKPYAVLLFDEFEKAHRDISALLLQVLDEGYLT 188
Query: 102 DSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRI----KSLVTEEL 157
D++G VDF+NT++++TSN+G+ ++ +G D + K+++ I + V + +
Sbjct: 189 DAQGHKVDFRNTIIVLTSNLGADIL------VGADPLHPYKETADGEIDPDVRKAVMDVV 350
Query: 158 KQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKT---------KEIDLSVT 208
+ PEFLNR+D IVF++L +++I DI + V D + L+
Sbjct: 351 AANYAPEFLNRIDSFIVFKRLALEALRDIVDISSRRVADSPRR*THLP*RT*SRS*LACG 530
Query: 209 ERFRDRVVEEGYDPSYGARPLRR 231
R R +V E P++ R +R
Sbjct: 531 ARLRPQVRSEAAKPAHHQRDWKR 599
Score = 61.6 bits (148), Expect = 3e-10
Identities = 33/85 (38%), Positives = 48/85 (55%), Gaps = 2/85 (2%)
Frame = +1
Query: 186 IADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKM 245
++ L E+ RL K I L V RD + E GYDP +GARPL R I + + +A+K+
Sbjct: 436 LSTFRLGELQTRLDDKRISLDVPNHVRDWLAERGYDPKFGARPLNRLITNEIGNGLADKI 615
Query: 246 LAGEIKEGD--SVIIDADSDGKVIV 268
+ GE+K G+ +V I D G +V
Sbjct: 616 IRGELKMGETATVAIKEDKSGLEVV 690
>BF632925 similar to GP|15425870|gb| heat shock protein 78 {Candida
albicans}, partial (2%)
Length = 525
Score = 80.1 bits (196), Expect = 9e-16
Identities = 36/70 (51%), Positives = 53/70 (75%)
Frame = -3
Query: 201 KEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDA 260
+EID V+E +D V +EGYDP+YGA PLR+AI+ L+ + +AE +LA + KEGD+V ID
Sbjct: 520 EEIDFEVSESVKDLVCKEGYDPTYGASPLRKAIVNLIANPLAEALLAEKCKEGDTVFIDL 341
Query: 261 DSDGKVIVLN 270
D++G +V+N
Sbjct: 340 DANGNTLVIN 311
>TC84370 weakly similar to PIR|T39572|T39572 probable proteinase subunit -
fission yeast (Schizosaccharomyces pombe), partial (13%)
Length = 643
Score = 80.1 bits (196), Expect = 9e-16
Identities = 41/122 (33%), Positives = 77/122 (62%), Gaps = 2/122 (1%)
Frame = +3
Query: 150 KSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLK--TKEIDLSV 207
+ LV L+ PEFLNR++ ++F +LT+ E+++I DI L E+ RL+ +++ + V
Sbjct: 12 RELVMNALRNCSLPEFLNRINSTVIFNRLTRREIRKIVDIRLLEIQKRLEGNGRKVYIDV 191
Query: 208 TERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVI 267
+E +D + GY P+YGARPL R I + + + +A +L G +++G++ ++ +G++
Sbjct: 192 SEEAKDYLGNAGYSPAYGARPLSRLIEKEVLNKLAILILRGNVRDGENARVEL-INGRIT 368
Query: 268 VL 269
VL
Sbjct: 369 VL 374
>TC81012 similar to PIR|T51523|T51523 clpB heat shock protein-like -
Arabidopsis thaliana, partial (11%)
Length = 577
Score = 78.2 bits (191), Expect = 3e-15
Identities = 38/95 (40%), Positives = 62/95 (65%)
Frame = +3
Query: 167 NRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGA 226
NR+DE IVFR L + ++ I + L+ V R+ +++ + VTE + GYDPSYGA
Sbjct: 3 NRVDEYIVFRPLDRDQISSIVRLQLERVQKRVADRKMKIRVTEPAIQLLGSLGYDPSYGA 182
Query: 227 RPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 261
RP++R I + +E+ +A+ +L GE KE D+++ID +
Sbjct: 183 RPVKRVIQQNVENELAKGILRGEFKEEDTILIDTE 287
>BG448326 similar to PIR|T51523|T51 clpB heat shock protein-like -
Arabidopsis thaliana, partial (22%)
Length = 646
Score = 75.5 bits (184), Expect = 2e-14
Identities = 34/52 (65%), Positives = 43/52 (82%)
Frame = +2
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPP 52
SF+F GPTGVGK+ELA TLASY F +E A++ +DMSE ME+H S+LIG+PP
Sbjct: 491 SFMFMGPTGVGKTELAXTLASYMFNTEXALVXIDMSESMEKHAXSRLIGAPP 646
>TC78991 similar to GP|9759268|dbj|BAB09589.1 101 kDa heat shock protein;
HSP101-like protein {Arabidopsis thaliana}, partial
(13%)
Length = 1370
Score = 57.0 bits (136), Expect = 8e-09
Identities = 32/118 (27%), Positives = 59/118 (49%)
Frame = +2
Query: 3 IFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQ 62
+F+GP +GK +A L+ GS +I L + + G T +
Sbjct: 458 MFTGPDRIGKKRMAAALSELVSGSNPIVISLAQRRGDGDSNAHQ-------FRGKTVLDR 616
Query: 63 LTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
+ E +RR P++V++ ++I++A+ + + + +E GR DS GR + N + I+TSN
Sbjct: 617 IVETIRRNPHSVIMLEDIDEANTLLRGNIKRAMEQGRFPDSHGREISLGNVMFILTSN 790
>TC82374 similar to GP|21689765|gb|AAM67526.1 putative arsA-like protein
hASNA-I {Arabidopsis thaliana}, partial (97%)
Length = 1176
Score = 32.0 bits (71), Expect = 0.28
Identities = 36/190 (18%), Positives = 76/190 (39%), Gaps = 14/190 (7%)
Frame = +2
Query: 45 SKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSK 104
++L G+ PG ++ + V+ Y+V++FD H +LE G
Sbjct: 299 NELAGAIPGIDEAMSFAEMLKLVQTMDYSVIVFDTAPTGHTLRLLQFPSVLEKGL----- 463
Query: 105 GRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSL--VTEELKQYFR 162
+ + KN + + R G D+ E D+ +++ + V E++ F+
Sbjct: 464 AKVMSLKNKF--------GGLFSQMTRMFGMGDDFGE-DAILGKLEGMKDVIEQVNMQFK 616
Query: 163 ------------PEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTER 210
PEFL+ + + ++LTK E+ I+ + +FD + L +
Sbjct: 617 DPDMTTFVCVCIPEFLSLYETERLVQELTKFEIDTHNIIINQVIFDDEDVESKLLKARMK 796
Query: 211 FRDRVVEEGY 220
+ + +++ Y
Sbjct: 797 MQQKYLDQFY 826
>TC84522 similar to GP|13161354|dbj|BAB32945. putative calcium sensor
protein {Oryza sativa (japonica cultivar-group)},
partial (59%)
Length = 1078
Score = 31.6 bits (70), Expect = 0.36
Identities = 26/119 (21%), Positives = 52/119 (42%), Gaps = 17/119 (14%)
Frame = -1
Query: 94 ILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNR----- 148
+L++ R++ K + F + +I+ NVG+ ++ +KI + + +SYN
Sbjct: 682 VLQERRISTHKKFPIFFTDFTIIVLINVGNCLVNNSFKKIIRKIHFRFHQNSYNHLLHFL 503
Query: 149 ------IKSLVTEELKQYF------RPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVF 195
+ +V + K YF R E +D+ I L +K I + + K++F
Sbjct: 502 WLNKPCLSQVVELKCKIYFFIHRGVRMENTEGVDKFIKLDNTVSLFIKNIKNSIQKKIF 326
>TC80970 similar to SP|O24357|G6PC_SPIOL Glucose-6-phosphate 1-dehydrogenase
chloroplast precursor (EC 1.1.1.49) (G6PD). [Spinach],
partial (55%)
Length = 1289
Score = 30.8 bits (68), Expect = 0.62
Identities = 32/148 (21%), Positives = 62/148 (41%), Gaps = 6/148 (4%)
Frame = +1
Query: 84 HPDVFNMMLQILE-DGRLTDSKGRTVDFKNTLLIMTSNVGSSVIE----KGGRKIGFDLD 138
H ++N L+ D +L + +G + + L + N+ V+ K K G+
Sbjct: 535 HSGLYNSEEDFLDLDSKLKEKEGGRLSNRLFYLSIPPNIFVDVVRCASLKASSKNGWTRV 714
Query: 139 YDEKDSSYN-RIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDR 197
EK + S +T LKQY + + R+D + + L V ++++ + ++ R
Sbjct: 715 IVEKPFGRDSESSSELTRSLKQYLTEDQIFRIDHYLGKELVENLSVLRFSNLVFEPLWSR 894
Query: 198 LKTKEIDLSVTERFRDRVVEEGYDPSYG 225
+ + L +E F GY +YG
Sbjct: 895 NYIRNVQLIFSEDFGTE-GRGGYFDNYG 975
>TC77030 similar to GP|7406669|emb|CAB85628.1 putative ripening-related
protein {Vitis vinifera}, partial (51%)
Length = 753
Score = 30.0 bits (66), Expect = 1.1
Identities = 38/143 (26%), Positives = 64/143 (44%), Gaps = 9/143 (6%)
Frame = +3
Query: 65 EAVRRRPYTVVLFDEIEKAHP---DVFNMMLQILED---GRLTDSKGRTVDFKNTLLIMT 118
+ +R Y +LFD + +P + +LQ ++D +L + D N L
Sbjct: 180 QQAQRSKYDCLLFDLDDTLYPLRAGLAKSVLQNIKDYMVEKLGIDSSKIDDLSNLLY--- 350
Query: 119 SNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELK--QYFRPEFLNRLDEMIVFR 176
N G+++ G R IG+D DYDE S + L E LK R L+ ++F
Sbjct: 351 KNYGTTM--SGLRAIGYDFDYDEYHSFVH--GRLPYENLKPDPILRNLLLSLPYRKLIFT 518
Query: 177 QLTKLE-VKEIADIMLKEVFDRL 198
K+ VK ++ + L++ F+ L
Sbjct: 519 NADKVHAVKALSRLGLEDCFEGL 587
>AW691664 similar to PIR|T52061|T52 tRNA isopentenyltransferase (EC
2.5.1.8) [imported] - Arabidopsis thaliana, partial
(32%)
Length = 665
Score = 29.3 bits (64), Expect = 1.8
Identities = 12/21 (57%), Positives = 17/21 (80%)
Frame = +3
Query: 3 IFSGPTGVGKSELAKTLASYY 23
+ +GPTG GKS+LA LAS++
Sbjct: 72 VITGPTGSGKSKLAVDLASHF 134
>TC77029 similar to GP|7406669|emb|CAB85628.1 putative ripening-related
protein {Vitis vinifera}, partial (83%)
Length = 1287
Score = 29.3 bits (64), Expect = 1.8
Identities = 37/141 (26%), Positives = 63/141 (44%), Gaps = 9/141 (6%)
Frame = +1
Query: 65 EAVRRRPYTVVLFDEIEKAHP---DVFNMMLQILED---GRLTDSKGRTVDFKNTLLIMT 118
+ +R Y +LFD + +P + +LQ ++D +L + D N L
Sbjct: 175 QQAQRSKYDCLLFDLDDTLYPLRAGLAKSVLQNIKDYMVEKLGIDSSKIDDLSNLLY--- 345
Query: 119 SNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELK--QYFRPEFLNRLDEMIVFR 176
N G+++ G R IG+D DYDE S + L E LK R L+ ++F
Sbjct: 346 KNYGTTM--SGLRAIGYDFDYDEYHSFVH--GRLPYENLKPDPILRNLLLSLPYRKLIFT 513
Query: 177 QLTKLE-VKEIADIMLKEVFD 196
K+ VK ++ + L++ F+
Sbjct: 514 NADKVHAVKALSRLGLEDCFE 576
>TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, complete
Length = 1793
Score = 28.9 bits (63), Expect = 2.3
Identities = 23/81 (28%), Positives = 36/81 (44%)
Frame = +2
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
+F+ GP G GKS LAK +A+ ++ + S+ VSK +G V
Sbjct: 683 AFLLYGPPGTGKSYLAKAVAT---EADSTFFSVSSSDL-----VSKWMGESEKLV----- 823
Query: 61 GQLTEAVRRRPYTVVLFDEIE 81
L E R +++ DEI+
Sbjct: 824 SNLFEMARESAPSIIFVDEID 886
>AW774182 homologue to PIR|F84674|F84 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, partial (27%)
Length = 487
Score = 28.9 bits (63), Expect = 2.3
Identities = 23/81 (28%), Positives = 36/81 (44%)
Frame = -1
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
+F+ GP G GKS LAK +A+ ++ + S+ VSK +G V
Sbjct: 247 AFLLYGPPGTGKSYLAKAVAT---KADSTFFSVSSSDL-----VSKWMGESEKLV----- 107
Query: 61 GQLTEAVRRRPYTVVLFDEIE 81
L E R +++ DEI+
Sbjct: 106 SNLFEMARESAPSIIFVDEID 44
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.137 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,672,941
Number of Sequences: 36976
Number of extensions: 56061
Number of successful extensions: 321
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 316
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 317
length of query: 284
length of database: 9,014,727
effective HSP length: 95
effective length of query: 189
effective length of database: 5,502,007
effective search space: 1039879323
effective search space used: 1039879323
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0266.20


