

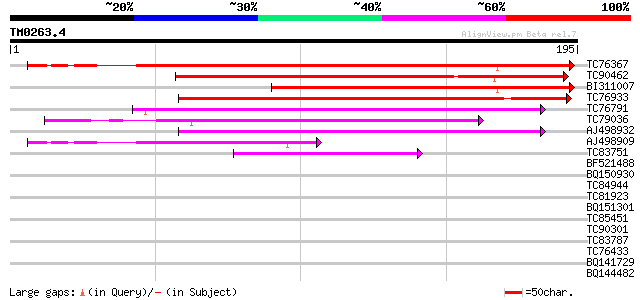
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0263.4
(195 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76367 similar to SP|P29530|OLE1_SOYBN P24 oleosin isoform A (P... 196 4e-51
TC90462 weakly similar to SP|P29530|OLE1_SOYBN P24 oleosin isofo... 168 1e-42
BI311007 similar to PIR|S17935|S179 oleosin (clone P24/89) - soy... 142 6e-35
TC76933 weakly similar to PIR|JC5703|JC5703 15.5K oleosin - orie... 112 9e-26
TC76791 similar to GP|21311555|gb|AAM46778.1 15.8 kDa oleosin {T... 100 5e-22
TC79036 similar to PIR|S51940|S51940 oleosin - almond, partial (... 84 5e-17
AJ498932 weakly similar to GP|9294318|dbj gene_id:K24M9.6~unknow... 81 2e-16
AJ498909 similar to SP|P29531|OLE2_ P24 oleosin isoform B (P91).... 65 2e-11
TC83751 weakly similar to GP|21311555|gb|AAM46778.1 15.8 kDa ole... 50 7e-07
BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila m... 30 0.59
BQ150930 similar to SP|O70133|DDX9 ATP-dependent RNA helicase A ... 29 1.0
TC84944 similar to GP|10177507|dbj|BAB10901. serine protease-lik... 28 1.7
TC81923 GP|21667854|gb|AAM74155.1 cyclin T1 protein {Capra hircu... 28 2.3
BQ151301 similar to PIR|S34666|S346 glycine-rich protein - commo... 24 2.8
TC85451 similar to PIR|T06239|T06239 probable glutathione transf... 28 3.0
TC90301 similar to GP|14194175|gb|AAK56282.1 AT5g58470/mqj2_60 {... 27 5.0
TC83787 similar to GP|9757890|dbj|BAB08397.1 phosphoribosylanthr... 27 5.0
TC76433 weakly similar to GP|15810483|gb|AAL07129.1 unknown prot... 27 5.0
BQ141729 homologue to GP|21629369|gb L712.4 {Leishmania major}, ... 27 5.0
BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 27 6.6
>TC76367 similar to SP|P29530|OLE1_SOYBN P24 oleosin isoform A (P89).
[Soybean] {Glycine max}, partial (50%)
Length = 909
Score = 196 bits (499), Expect = 4e-51
Identities = 111/195 (56%), Positives = 130/195 (65%), Gaps = 7/195 (3%)
Frame = +2
Query: 7 QVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSASQIIA 66
Q+ VHTT T RY+T PP YE GGGG + PS SQ++A
Sbjct: 95 QIQVHTT-TRRYDT--ITPPLHYE-------------GGGGVATHHYSDNKSPSTSQVLA 226
Query: 67 VAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGAF 126
V G+P+GG LL LAG+SL+ +L GLAV TPL ILFSPVIVPA + IGLAVAG LTSGAF
Sbjct: 227 VITGLPVGGILLALAGLSLLGTLTGLAVTTPLFILFSPVIVPATIVIGLAVAGFLTSGAF 406
Query: 127 GLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYV-------GQKTKEVGHDIQT 179
GLT L SFSW+MNYIR+T+G VPEQL+ AK MAD+ YV GQKTKEVG DIQ+
Sbjct: 407 GLTALSSFSWVMNYIRQTQGTVPEQLESAKQGMADVVGYVGQKTKEAGQKTKEVGQDIQS 586
Query: 180 KAHEAKRTPITTHDT 194
KA +AK+T TT T
Sbjct: 587 KAQDAKKTSTTTTAT 631
>TC90462 weakly similar to SP|P29530|OLE1_SOYBN P24 oleosin isoform A (P89).
[Soybean] {Glycine max}, partial (46%)
Length = 591
Score = 168 bits (425), Expect = 1e-42
Identities = 92/142 (64%), Positives = 103/142 (71%), Gaps = 7/142 (4%)
Frame = +3
Query: 58 GPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAV 117
G SASQI+A G+ +GGTLLLLA S ASLIGLA+ TPL ILFSPV++PA +TIGLAV
Sbjct: 30 GSSASQILASLGGLFLGGTLLLLASFSFFASLIGLAIMTPLFILFSPVLIPAALTIGLAV 209
Query: 118 AGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADY-------VGQKT 170
AGILT+ A GLTGLMS SW YIR+ VP Q+D K RMAD+ DY VGQKT
Sbjct: 210 AGILTADACGLTGLMSLSWTARYIRDLHAAVPGQMD-VKGRMADVVDYVGQKTKDVGQKT 386
Query: 171 KEVGHDIQTKAHEAKRTPITTH 192
KEVG DIQTKAHEAKRT H
Sbjct: 387 KEVGQDIQTKAHEAKRTT*INH 452
>BI311007 similar to PIR|S17935|S179 oleosin (clone P24/89) - soybean
(fragment), partial (53%)
Length = 760
Score = 142 bits (359), Expect = 6e-35
Identities = 76/111 (68%), Positives = 85/111 (76%), Gaps = 7/111 (6%)
Frame = +3
Query: 91 GLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPE 150
GLAV TPL ILFSPVIVPA + IGLAVAG LTSGAFGLT L SFSW+MNYIR+T+G VPE
Sbjct: 3 GLAVTTPLFILFSPVIVPATIVIGLAVAGFLTSGAFGLTALSSFSWVMNYIRQTQGTVPE 182
Query: 151 QLDYAKNRMADMADYV-------GQKTKEVGHDIQTKAHEAKRTPITTHDT 194
QL+ AK MAD+ YV GQKTKEVG DIQ+KA +AK+T TT T
Sbjct: 183 QLESAKQGMADVVGYVGQKTKEAGQKTKEVGQDIQSKAQDAKKTSTTTTAT 335
>TC76933 weakly similar to PIR|JC5703|JC5703 15.5K oleosin - oriental
sesame, partial (60%)
Length = 907
Score = 112 bits (280), Expect = 9e-26
Identities = 57/135 (42%), Positives = 86/135 (63%)
Frame = +2
Query: 59 PSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVA 118
PS SQ+I +A VP G TLL+LAG++L A+++GLAV TPL I FSP+++ A + IGLA+A
Sbjct: 161 PSTSQLIVLATLVPFGATLLILAGLTLTATVVGLAVTTPLFIFFSPILLAAAVVIGLAIA 340
Query: 119 GILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHDIQ 178
G LTSGAFG+T L SF+W+ +Y+R +R + + + + +A + +T +GH+ Q
Sbjct: 341 GFLTSGAFGITSLSSFAWVASYLRRSRFLEKVNVKHHHHAIAKPPRFDADET--LGHESQ 514
Query: 179 TKAHEAKRTPITTHD 193
+ R D
Sbjct: 515 INLEDRDRVESMAQD 559
>TC76791 similar to GP|21311555|gb|AAM46778.1 15.8 kDa oleosin {Theobroma
cacao}, partial (64%)
Length = 933
Score = 100 bits (248), Expect = 5e-22
Identities = 50/144 (34%), Positives = 80/144 (54%), Gaps = 2/144 (1%)
Frame = +1
Query: 43 YGG--GGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLI 100
YGG GG + + + Q + IG TLL L+G+ L+ ++IGL + TPLL+
Sbjct: 154 YGGSYGGSPYDTNINNNNQPSRQTVKFITAATIGVTLLFLSGLILVGTVIGLIIATPLLV 333
Query: 101 LFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMA 160
+FSP++VPA + + L G + SG G+ + + SW+ NY+ R + LDYAK +
Sbjct: 334 IFSPILVPAAIVLSLIAGGFMFSGGCGVAAIAALSWIYNYVSGNRPAGTDTLDYAKGLIT 513
Query: 161 DMADYVGQKTKEVGHDIQTKAHEA 184
D A V ++ K+ G+ Q +A +A
Sbjct: 514 DKARDVKERAKDYGNYAQGRAQDA 585
>TC79036 similar to PIR|S51940|S51940 oleosin - almond, partial (67%)
Length = 704
Score = 83.6 bits (205), Expect = 5e-17
Identities = 49/153 (32%), Positives = 81/153 (52%), Gaps = 2/153 (1%)
Frame = +1
Query: 13 TATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSA--SQIIAVAAG 70
T T+ +TGG P R +N Q S ++ PS+ +Q++ +
Sbjct: 37 TLTNMADTGGHYQPLR------GYNQQ-----------HSTTTQQQPSSKLTQLLKSSTA 165
Query: 71 VPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGAFGLTG 130
V GG+LL+L+G+ L ++I L + TPL +LFSPV+VPAV+T+ L G SG FG+
Sbjct: 166 VTAGGSLLILSGLVLAGTVIALTIATPLFLLFSPVLVPAVITVALLTLGFFVSGGFGVAA 345
Query: 131 LMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
+ +W+ Y+ +QLD A++++ + A
Sbjct: 346 ITVLAWIYRYVTGKHPPGADQLDTARHKLMNKA 444
>AJ498932 weakly similar to GP|9294318|dbj gene_id:K24M9.6~unknown protein
{Arabidopsis thaliana}, partial (62%)
Length = 565
Score = 81.3 bits (199), Expect = 2e-16
Identities = 41/126 (32%), Positives = 72/126 (56%)
Frame = +2
Query: 59 PSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVA 118
P+++Q+ + + G LLL G+++ A+++ L F+PL+I+ SP+ VPA + L A
Sbjct: 98 PNSTQLAGLLTLLVTGSIFLLLTGLTVAATVVSLIFFSPLIIVSSPIWVPAGTFLFLLAA 277
Query: 119 GILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHDIQ 178
G L+ FG+ + + SW Y R +++DYA+NR+ D A +V +E G +Q
Sbjct: 278 GFLSMCGFGVVAVAASSWFYRYFRGLHPPGSDRVDYARNRLYDTASHVKDYAREYGGYLQ 457
Query: 179 TKAHEA 184
+K +A
Sbjct: 458 SKVKDA 475
>AJ498909 similar to SP|P29531|OLE2_ P24 oleosin isoform B (P91). [Soybean]
{Glycine max}, partial (21%)
Length = 464
Score = 64.7 bits (156), Expect = 2e-11
Identities = 41/104 (39%), Positives = 56/104 (53%), Gaps = 3/104 (2%)
Frame = +2
Query: 7 QVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSASQIIA 66
Q+ VHTT T RY+T PP YE GGGG + PS SQ++A
Sbjct: 86 QIQVHTT-TRRYDT--ITPPLHYE-------------GGGGVATHHYSDNKSPSTSQVLA 217
Query: 67 VAAGVPIGGTLLLLAGISLIASLIGLAV---FTPLLILFSPVIV 107
V G+P+GG LL LAG+SL+ +L GLA+ F + +F ++V
Sbjct: 218 VITGLPVGGILLALAGLSLLGTLTGLALSLCFVRKVCVFCDLVV 349
>TC83751 weakly similar to GP|21311555|gb|AAM46778.1 15.8 kDa oleosin
{Theobroma cacao}, partial (29%)
Length = 860
Score = 49.7 bits (117), Expect = 7e-07
Identities = 19/65 (29%), Positives = 37/65 (56%)
Frame = +1
Query: 78 LLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWL 137
L L ++ + +++G +F P++IL SP++VP + + + G L + FG+ L SW+
Sbjct: 133 LFLTCLTFVVTIMGFILFAPMIILLSPILVPVFAVLFVFIVGFLFTCGFGIVVLAMLSWI 312
Query: 138 MNYIR 142
Y +
Sbjct: 313 FRYFK 327
>BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila
melanogaster}, partial (84%)
Length = 470
Score = 30.0 bits (66), Expect = 0.59
Identities = 24/64 (37%), Positives = 28/64 (43%)
Frame = +2
Query: 21 GGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLL 80
GG P + GG F +P R GGGGGG S+ SAS A G GG+
Sbjct: 239 GGGRPYGGFGGGG--FGGRPFRGGGGGGGGSA-------SASASANAAGGGFRGGSAAAS 391
Query: 81 AGIS 84
A S
Sbjct: 392 AAAS 403
>BQ150930 similar to SP|O70133|DDX9 ATP-dependent RNA helicase A (Nuclear DNA
helicase II) (NDH II) (DEAD-box protein 9) (mHEL-5).,
partial (1%)
Length = 826
Score = 29.3 bits (64), Expect = 1.0
Identities = 18/49 (36%), Positives = 22/49 (44%)
Frame = -3
Query: 5 QQQVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSL 53
++QV HT T YET NP G+NF N + G G SL
Sbjct: 593 ERQVETHTYITQYYET---NPADEITWIGVNFENGDRERGTSMGTEESL 456
>TC84944 similar to GP|10177507|dbj|BAB10901. serine protease-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 960
Score = 28.5 bits (62), Expect = 1.7
Identities = 23/89 (25%), Positives = 45/89 (49%), Gaps = 9/89 (10%)
Frame = -2
Query: 27 QRYEAGGIN-----FNNQPQRYG----GGGGGFSSLFPERGPSASQIIAVAAGVPIGGTL 77
+R AGG + F +P+R GG SS FP S+ + ++A V + +
Sbjct: 374 RRRPAGGFSGIAGCFLGRPRRPRPS*EGGPLSMSSTFPTEKSSSIIVEGLSATVVVAVVV 195
Query: 78 LLLAGISLIASLIGLAVFTPLLILFSPVI 106
+++ +++++ + G+ PLL L SP++
Sbjct: 194 VVV--VAVVSRVSGVLGLRPLLRLLSPIV 114
>TC81923 GP|21667854|gb|AAM74155.1 cyclin T1 protein {Capra hircus}, partial
(1%)
Length = 839
Score = 28.1 bits (61), Expect = 2.3
Identities = 16/42 (38%), Positives = 20/42 (47%)
Frame = -1
Query: 18 YETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGP 59
YE+GG Y GG +Q YG GGGF + + GP
Sbjct: 818 YESGG------YGGGGGVMMHQGHVYGSVGGGFHQVMGKGGP 711
>BQ151301 similar to PIR|S34666|S346 glycine-rich protein - common tobacco,
partial (22%)
Length = 530
Score = 24.3 bits (51), Expect(2) = 2.8
Identities = 14/30 (46%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Frame = -2
Query: 21 GGANPPQRYEAGGINFNN-QPQRYGGGGGG 49
GG P R GG +N +P+ GGGGGG
Sbjct: 211 GGWVPCGRGGGGGRGGHNYRPRLMGGGGGG 122
Score = 21.9 bits (45), Expect(2) = 2.8
Identities = 10/15 (66%), Positives = 10/15 (66%)
Frame = -2
Query: 45 GGGGGFSSLFPERGP 59
GGGGGF S FP P
Sbjct: 91 GGGGGFGS-FPPPSP 50
>TC85451 similar to PIR|T06239|T06239 probable glutathione transferase (EC
2.5.1.18) 2 4-D inducible - soybean, complete
Length = 1155
Score = 27.7 bits (60), Expect = 3.0
Identities = 22/90 (24%), Positives = 42/90 (46%), Gaps = 2/90 (2%)
Frame = +3
Query: 96 TPLLILFSPVI--VPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLD 153
+PLL+ +PV +P ++ G ++ L++ ++ E ++P
Sbjct: 270 SPLLLQMNPVHKKIPVLIHNGKSIC----------ESLIAVQYIDEVWNEKSPLLPSD-P 416
Query: 154 YAKNRMADMADYVGQKTKEVGHDIQTKAHE 183
Y +++ ADYV +K EVG ++ TK E
Sbjct: 417 YQRSQARFWADYVDKKIYEVGRNLWTKKGE 506
>TC90301 similar to GP|14194175|gb|AAK56282.1 AT5g58470/mqj2_60 {Arabidopsis
thaliana}, partial (46%)
Length = 1222
Score = 26.9 bits (58), Expect = 5.0
Identities = 13/33 (39%), Positives = 15/33 (45%)
Frame = +1
Query: 18 YETGGANPPQRYEAGGINFNNQPQRYGGGGGGF 50
Y T PP Y G N+ P YGG GG+
Sbjct: 223 YGTDAVPPPTSYTGGP---NSYPPSYGGNTGGY 312
>TC83787 similar to GP|9757890|dbj|BAB08397.1 phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(11%)
Length = 1008
Score = 26.9 bits (58), Expect = 5.0
Identities = 15/38 (39%), Positives = 19/38 (49%)
Frame = -1
Query: 21 GGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERG 58
GG P + GG F++ GGGG SS FP+ G
Sbjct: 321 GGGAPDG*SDGGGSTFSSSD-----GGGGDSSTFPDGG 223
>TC76433 weakly similar to GP|15810483|gb|AAL07129.1 unknown protein
{Arabidopsis thaliana}, partial (61%)
Length = 2810
Score = 26.9 bits (58), Expect = 5.0
Identities = 25/84 (29%), Positives = 36/84 (42%), Gaps = 3/84 (3%)
Frame = +1
Query: 48 GGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIV 107
GGFS +FP+ SA I+A A P L + L +G+ + P L L + +
Sbjct: 259 GGFSGIFPDSSDSA-YILAAATSTP---NAALWCDVQLTKDAVGICL--PNLNLLNSTNI 420
Query: 108 PAVM---TIGLAVAGILTSGAFGL 128
V T V G T+G F +
Sbjct: 421 FGVFPNKTTSYLVNGESTAGYFSV 492
>BQ141729 homologue to GP|21629369|gb L712.4 {Leishmania major}, partial (1%)
Length = 1078
Score = 26.9 bits (58), Expect = 5.0
Identities = 19/59 (32%), Positives = 25/59 (42%), Gaps = 13/59 (22%)
Frame = -1
Query: 4 PQQQVH-----VHTTATHRYETGGANPPQRYEA--------GGINFNNQPQRYGGGGGG 49
P+ Q H VHT ++ +TG P +R GG + R GGGGGG
Sbjct: 190 PRPQTHSAHPCVHTHVRNKDKTGVPYPERRTRTNGAARAPTGGARTSGARWRGGGGGGG 14
>BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (16%)
Length = 993
Score = 26.6 bits (57), Expect = 6.6
Identities = 16/37 (43%), Positives = 17/37 (45%)
Frame = -2
Query: 21 GGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPER 57
GG PP R F P GGGG FS LFP +
Sbjct: 626 GGKGPPPRPF-----FGLPPPF*KGGGGDFSKLFPRQ 531
Score = 26.2 bits (56), Expect = 8.6
Identities = 16/41 (39%), Positives = 20/41 (48%)
Frame = -3
Query: 22 GANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSAS 62
G PP GGI N+ P + GGG S +RGP +S
Sbjct: 592 GFPPPFERGGGGIFQNSFPAKPTGGGPPVSP*KKKRGPFSS 470
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,167,569
Number of Sequences: 36976
Number of extensions: 67392
Number of successful extensions: 623
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 567
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 612
length of query: 195
length of database: 9,014,727
effective HSP length: 91
effective length of query: 104
effective length of database: 5,649,911
effective search space: 587590744
effective search space used: 587590744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0263.4


