

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
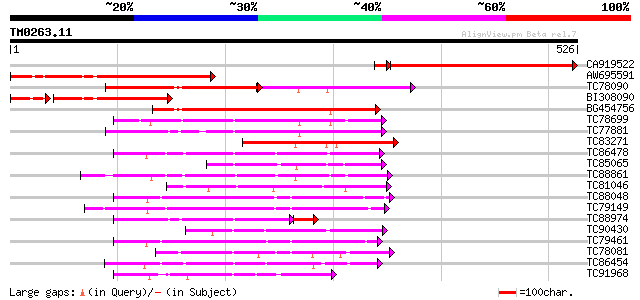
Query= TM0263.11
(526 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA919522 similar to PIR|S53804|S53 protein kinase NPK2 (EC 2.7.1... 319 2e-88
AW695591 similar to PIR|T51338|T51 mitogen-activated protein kin... 317 9e-87
TC78090 GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago sa... 135 7e-60
BI308090 similar to GP|21655213|gb mitogen-activated protein kin... 192 1e-57
BG454756 homologue to GP|12718822|db NQK1 MAPKK {Nicotiana tabac... 202 2e-52
TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase ... 171 6e-43
TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase ki... 163 1e-40
TC83271 similar to GP|15528441|emb|CAC69138. MAP kinase kinase {... 160 1e-39
TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 prote... 141 5e-34
TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated p... 117 1e-26
TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative pro... 117 1e-26
TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein ki... 110 2e-24
TC88048 similar to GP|20302596|dbj|BAB91125. Ser/Thr kinase {Ara... 107 1e-23
TC79149 similar to PIR|T46059|T46059 MAP kinase - Arabidopsis th... 105 3e-23
TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Bra... 99 7e-23
TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 103 1e-22
TC79461 similar to GP|9758254|dbj|BAB08753.1 contains similarity... 96 3e-20
TC78081 similar to GP|16904226|gb|AAL30820.1 calcium/calmodulin-... 96 3e-20
TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine... 93 2e-19
TC91968 similar to PIR|T02550|T02550 NPK1-related protein kinase... 93 2e-19
>CA919522 similar to PIR|S53804|S53 protein kinase NPK2 (EC 2.7.1.-) - common
tobacco, partial (33%)
Length = 799
Score = 319 bits (817), Expect(2) = 2e-88
Identities = 151/173 (87%), Positives = 161/173 (92%)
Frame = -1
Query: 354 EVDLAGYVRSVFDPTQRMKDLADMLTIHYYLLFDGPVDLWQNARNLYSESSIFSFSGKQH 413
+VDL G RSVFDPTQRMKDLADMLTIHYYLLFDGP D WQ+ RN Y+E+SIFSFSGKQH
Sbjct: 751 KVDLPGLFRSVFDPTQRMKDLADMLTIHYYLLFDGPDDSWQHTRNFYNENSIFSFSGKQH 572
Query: 414 VGPNNIFKTLSSIRCTLVGEWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIVGNQFLIC 473
+GPNNIF TLSSIR TL+G+WPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFI+GNQFLIC
Sbjct: 571 IGPNNIFTTLSSIRTTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIIGNQFLIC 392
Query: 474 GDGIQVEGLPNFKDLDIDISSKRMGTFHEQFIVEPTTHIGCYTIVNQELYINQ 526
GDG+QVEGLPNFKDL IDISSKRMGTFHEQFIVEPT+ IGCYTIV QELYINQ
Sbjct: 391 GDGVQVEGLPNFKDLGIDISSKRMGTFHEQFIVEPTSQIGCYTIVKQELYINQ 233
Score = 25.8 bits (55), Expect(2) = 2e-88
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = -3
Query: 339 EQLLFHPFITKYETVE 354
EQL PFITKYET +
Sbjct: 797 EQLFCTPFITKYETAK 750
>AW695591 similar to PIR|T51338|T51 mitogen-activated protein kinase kinase
(EC 2.7.1.-) 3 [imported] - Arabidopsis thaliana,
partial (34%)
Length = 649
Score = 317 bits (811), Expect = 9e-87
Identities = 165/191 (86%), Positives = 175/191 (91%)
Frame = +2
Query: 1 MAGLEELRKKLAPLFDAEKGFGFSTSSTLDPCDDSYTLSDGGTVNLLSRSYGVYNINELG 60
M+GLEELRKKL PLFDAE GF S++S+ DP +DSYT SDGGTVNLLSRSYGVYNINELG
Sbjct: 98 MSGLEELRKKLTPLFDAENGF--SSTSSFDP-NDSYTFSDGGTVNLLSRSYGVYNINELG 268
Query: 61 LQKCSSSSRSVDESHDSDHSEKTYRCGSREMRIFGAIGSGASSVVQRAIHIPKHRIIALK 120
LQKCS+S RSVDES D+ EKTYRCGS EMRIFGAIGSGASSVVQRA+HIP HR+IALK
Sbjct: 269 LQKCSTS-RSVDESDDN---EKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALK 436
Query: 121 KINIFEKEKRQQLLTEIRTLCEAPCYQGLVEFHGAFYTPDSGQISIALEYMDGGSLADIL 180
KINIFEKEKRQQLLTEIRTLCEAPCY+GLVEFHGAFYTPDSGQISIALEYMDGGSLADIL
Sbjct: 437 KINIFEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADIL 616
Query: 181 RKHRRIPEPIL 191
R HR IPEPIL
Sbjct: 617 RMHRTIPEPIL 649
>TC78090 GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago sativa subsp.
x varia}, complete
Length = 1677
Score = 135 bits (339), Expect(2) = 7e-60
Identities = 71/155 (45%), Positives = 92/155 (58%), Gaps = 8/155 (5%)
Frame = +3
Query: 230 KITDFGISAGLESSVAMCATFVGTVTYMSPERIRNES--YSYPADIWSLGLALLECGTGE 287
++ DFG+SA +ES+ TF+GT YMSPERI Y+Y +DIWSLGL LLEC G
Sbjct: 840 RLPDFGVSAIMESTSGQANTFIGTYNYMSPERINGSQRGYNYKSDIWSLGLILLECAMGR 1019
Query: 288 FPYTAN------EGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQL 341
FPYT E L+ I+D P PS + FS EFCSFI ACLQKDP R +A++L
Sbjct: 1020FPYTPPDQSERWESIFELIETIVDKPPPSAPSEQFSSEFCSFISACLQKDPGSRLSAQEL 1199
Query: 342 LFHPFITKYETVEVDLAGYVRSVFDPTQRMKDLAD 376
+ PFI+ Y+ + VDL+ Y P + + D
Sbjct: 1200MELPFISMYDDLHVDLSAYFSDAGSPLATL*KMID 1304
Score = 114 bits (285), Expect(2) = 7e-60
Identities = 66/146 (45%), Positives = 95/146 (64%), Gaps = 1/146 (0%)
Frame = +2
Query: 90 EMRIFGAIGSGASSVVQRAIHIPKHRIIALKKINI-FEKEKRQQLLTEIRTLCEAPCYQG 148
++ I +G G VVQ H ++ ALK I + E+ R+Q+ E++ A C
Sbjct: 425 DIDIVKVVGKGNGGVVQLVQHKWTNQFFALKIIQMNIEESVRKQIAKELKINQAAQCPYV 604
Query: 149 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGV 208
+V + +FY D+G ISI LEYMDGGS+AD+L+K + IPEP LS++ +++L GL YLH
Sbjct: 605 VVCYQ-SFY--DNGVISIILEYMDGGSMADLLKKVKTIPEPYLSAICKQVLKGLIYLHHE 775
Query: 209 RHLVHRDIKPANLLVNLKGEPKITDF 234
RH++HRD+KP+NLL+N GE KIT F
Sbjct: 776 RHIIHRDLKPSNLLINHTGEVKIT*F 853
>BI308090 similar to GP|21655213|gb mitogen-activated protein kinase kinase
{Suaeda maritima subsp. salsa}, partial (23%)
Length = 482
Score = 192 bits (488), Expect(2) = 1e-57
Identities = 99/111 (89%), Positives = 104/111 (93%)
Frame = +3
Query: 41 GGTVNLLSRSYGVYNINELGLQKCSSSSRSVDESHDSDHSEKTYRCGSREMRIFGAIGSG 100
GGTVNLLSRSYGVYNINELGLQKCS+S RSVDES D+ EKTYRCGS EMRIFGAIGSG
Sbjct: 162 GGTVNLLSRSYGVYNINELGLQKCSTS-RSVDESDDN---EKTYRCGSHEMRIFGAIGSG 329
Query: 101 ASSVVQRAIHIPKHRIIALKKINIFEKEKRQQLLTEIRTLCEAPCYQGLVE 151
ASSVVQRA+HIP HR+IALKKINIFEKEKRQQLLTEIRTLCEAPCY+GLVE
Sbjct: 330 ASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTLCEAPCYEGLVE 482
Score = 49.7 bits (117), Expect(2) = 1e-57
Identities = 26/38 (68%), Positives = 32/38 (83%)
Frame = +1
Query: 1 MAGLEELRKKLAPLFDAEKGFGFSTSSTLDPCDDSYTL 38
M+GLEELRKKL PLFDAE GFS++S+ DP +DSYT+
Sbjct: 58 MSGLEELRKKLTPLFDAEN--GFSSTSSFDP-NDSYTM 162
>BG454756 homologue to GP|12718822|db NQK1 MAPKK {Nicotiana tabacum}, partial
(60%)
Length = 648
Score = 202 bits (515), Expect = 2e-52
Identities = 105/218 (48%), Positives = 143/218 (65%), Gaps = 6/218 (2%)
Frame = +2
Query: 133 LLTEIRTLCEAPCYQGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILS 192
++ E++ + C +V +H +FY ++G IS+ LEYMD GSL D++R+ I EP L+
Sbjct: 2 IVQELKINQASQCPHVVVCYH-SFY--NNGVISLVLEYMDRGSLVDVIRQVNTILEPYLA 172
Query: 193 SMFQKLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVG 252
+ +++L GL YLH RH++HRDIKP+NLLVN KGE KITDFG+SA L S++ TFVG
Sbjct: 173 VVCKQVLQGLVYLHNERHVIHRDIKPSNLLVNHKGEVKITDFGVSAMLASTMGQRDTFVG 352
Query: 253 TVTYMSPERIRNESYSYPADIWSLGLALLECGTGEFPYTANEGP------VNLMLQILDD 306
T YMSPERI +Y Y DIWSLG+ +LEC G FPY +E L+ I++
Sbjct: 353 TYNYMSPERISGSTYDYSCDIWSLGMVVLECAIGRFPYIQSEDQQAWPSFYELLQAIVES 532
Query: 307 PSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFH 344
P PS FSPEFCSF+ +C++KDP R T+ +LL H
Sbjct: 533 PPPSAPPDQFSPEFCSFVSSCIKKDPRERSTSLELLDH 646
>TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase T9L24.32
[imported] - Arabidopsis thaliana, partial (71%)
Length = 1119
Score = 171 bits (433), Expect = 6e-43
Identities = 108/264 (40%), Positives = 154/264 (57%), Gaps = 11/264 (4%)
Frame = +3
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKINIFEKEK--RQQLLTEIRTLCEAPCYQGLVEFHG 154
+G G V + H I ALK IN ++ + R++ LTE+ L A +V++HG
Sbjct: 219 LGHGNGGTVYKVRHKLTSIIYALK-INHYDSDPTTRRRALTEVNILRRATDCTNVVKYHG 395
Query: 155 AFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHLVHR 214
+F P +G + I +EYMD GSL L+ E LS++ + +L+GL+YLH R++ HR
Sbjct: 396 SFEKP-TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHA-RNIAHR 569
Query: 215 DIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNESY-----SY 269
DIKP+N+LVN+K E KI DFG+S + ++ C ++VGT YMSPER E Y +
Sbjct: 570 DIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGF 749
Query: 270 PADIWSLGLALLECGTGEFPY-TANEGP--VNLMLQI-LDDPSPSPSKQTFSPEFCSFID 325
ADIWSLGL L E G FP+ + + P +LM I DP P +T S EF +F++
Sbjct: 750 SADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLP--ETASSEFRNFVE 923
Query: 326 ACLQKDPDVRPTAEQLLFHPFITK 349
CL+K+ R +A QLL HPF+ K
Sbjct: 924 CCLKKESGERWSAAQLLTHPFLCK 995
>TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase kinsae
{Medicago sativa subsp. x varia}, complete
Length = 1546
Score = 163 bits (413), Expect = 1e-40
Identities = 103/268 (38%), Positives = 152/268 (56%), Gaps = 8/268 (2%)
Frame = +2
Query: 90 EMRIFGAIGSGASSVVQRAIHIPKHRIIALKKI-NIFEKEKRQQLLTEIRTLCEAPCYQG 148
E+ IGSG+ V + +H R ALK I E+ R+Q+ EI+ L +
Sbjct: 464 ELERLNRIGSGSGGTVYKVVHRINGRAYALKVIYGHHEESVRRQIHREIQILRDVDDVN- 640
Query: 149 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRKHRRIP-EPILSSMFQKLLHGLSYLHG 207
+V+ H + + +I + LEYMDGGSL + + IP E L+ + +++L GL+YLH
Sbjct: 641 VVKCHEMY--DHNAEIQVLLEYMDGGSL-----EGKHIPQENQLADVARQILRGLAYLHR 799
Query: 208 VRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNESY 267
RH+VHRDIKP+NLL+N + + KI DFG+ L ++ C + VGT+ YMSPERI +
Sbjct: 800 -RHIVHRDIKPSNLLINSRKQVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDIN 976
Query: 268 -----SYPADIWSLGLALLECGTGEFPY-TANEGPVNLMLQILDDPSPSPSKQTFSPEFC 321
+Y DIWSLG+++LE G FP+ +G ++ + P + T SPEF
Sbjct: 977 DGQYDAYAGDIWSLGVSILEFYMGRFPFAVGRQGDWASLMCAICMSQPPEAPTTASPEFR 1156
Query: 322 SFIDACLQKDPDVRPTAEQLLFHPFITK 349
F+ CLQ+DP R TA +LL HPF+ +
Sbjct: 1157DFVSRCLQRDPSRRWTASRLLSHPFLVR 1240
>TC83271 similar to GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago
sativa subsp. x varia}, partial (42%)
Length = 683
Score = 160 bits (405), Expect = 1e-39
Identities = 82/154 (53%), Positives = 105/154 (67%), Gaps = 10/154 (6%)
Frame = +1
Query: 217 KPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIR----NESYSYPAD 272
KP+N+L+N +GE KITDFG+SA +ES+ TF+GT YMSPERI+ + Y+Y +D
Sbjct: 10 KPSNILINHRGEVKITDFGVSAIMESTSGQANTFIGTYNYMSPERIKASDSEQGYNYKSD 189
Query: 273 IWSLGLALLECGTGEFPYTA---NEGPVNLML---QILDDPSPSPSKQTFSPEFCSFIDA 326
IWSLGL LL+C TG+FPYT +EG NL L I++ PSPS SPEFCSFI A
Sbjct: 190 IWSLGLMLLKCATGKFPYTPPDNSEGWENLFLLIEAIVEKPSPSAPPDECSPEFCSFISA 369
Query: 327 CLQKDPDVRPTAEQLLFHPFITKYETVEVDLAGY 360
CLQK+P RP+ LL HPF+ Y+ + VDL+ Y
Sbjct: 370 CLQKNPRDRPSTRNLLRHPFVNMYDDLHVDLSDY 471
>TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 protein kinase
{Brassica napus}, partial (52%)
Length = 2266
Score = 141 bits (356), Expect = 5e-34
Identities = 90/257 (35%), Positives = 149/257 (57%), Gaps = 6/257 (2%)
Frame = +2
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKINIF-----EKEKRQQLLTEIRTLCEAPCYQGLVE 151
+G G V + ++ A+K++ + KE +QL EI L + + +V+
Sbjct: 989 LGRGTFGHVYLGFNSENGQMCAIKEVRVGCDDQNSKECLKQLHQEIDLLNQLS-HPNIVQ 1165
Query: 152 FHGAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHL 211
+ G+ +S +S+ LEY+ GGS+ +L+++ EP++ + ++++ GL+YLHG R+
Sbjct: 1166 YLGSELGEES--LSVYLEYVSGGSIHKLLQEYGPFKEPVIQNYTRQIVSGLAYLHG-RNT 1336
Query: 212 VHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRN-ESYSYP 270
VHRDIK AN+LV+ GE K+ DFG++ + S+ +M +F G+ +M+PE + N YS P
Sbjct: 1337 VHRDIKGANILVDPNGEIKLADFGMAKHITSAASM-LSFKGSPYWMAPEVVMNTNGYSLP 1513
Query: 271 ADIWSLGLALLECGTGEFPYTANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQK 330
DIWSLG L+E + P++ EG V + +I + + S + +FI CLQ+
Sbjct: 1514 VDIWSLGCTLIEMAASKPPWSQYEG-VAAIFKIGNSKDMPIIPEHLSNDAKNFIMLCLQR 1690
Query: 331 DPDVRPTAEQLLFHPFI 347
DP RPTA++LL HPFI
Sbjct: 1691 DPSARPTAQKLLEHPFI 1741
>TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated protein
kinase {Arabidopsis thaliana}, partial (26%)
Length = 789
Score = 117 bits (293), Expect = 1e-26
Identities = 71/170 (41%), Positives = 102/170 (59%), Gaps = 3/170 (1%)
Frame = +2
Query: 183 HRRIPEPILSSMFQKLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLE- 241
H ++ + +S+ +++LHGL YLH R++VHRDIK AN+LV+ G K+ DFG+S ++
Sbjct: 2 HEKLRDSQVSAYTRQILHGLKYLHD-RNIVHRDIKCANILVDANGSVKVADFGLS*AIKL 178
Query: 242 SSVAMCATFVGTVTYMSPERIRN--ESYSYPADIWSLGLALLECGTGEFPYTANEGPVNL 299
+ V C GT +M+PE +R + Y PADIWSLG +LE TG+ PY E ++
Sbjct: 179 NDVKSCQ---GTAFWMAPEVVRGKVKGYGLPADIWSLGCTVLEMLTGQVPYAPME-CISA 346
Query: 300 MLQILDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFITK 349
M +I P P T S + FI CL+ +PD RPTA QLL H F+ +
Sbjct: 347 MFRIGKGELP-PVPDTLSRDARDFILQCLKVNPDDRPTAAQLLDHKFVQR 493
>TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (34%)
Length = 1847
Score = 117 bits (292), Expect = 1e-26
Identities = 93/304 (30%), Positives = 158/304 (51%), Gaps = 14/304 (4%)
Frame = +1
Query: 66 SSSRSVDESHDSDHSEKTYRCGSREMRIFGAIGSGASSVVQRAIHIPKHRIIALKKINIF 125
+SS +++ S ++ HS K + + IG G+ V A ++ ALK++++
Sbjct: 967 NSSVTMNHSTENIHSMKGHWQKGK------LIGRGSFGSVYHATNLETGASCALKEVDLV 1128
Query: 126 EKEKR-----QQLLTEIRTLCEAPCYQGLVEFHGAFYTPDSGQISIALEYMDGGSLADIL 180
+ + +QL EIR L + + +VE++G+ D ++ I +EY+ GSL +
Sbjct: 1129 PDDPKSTDCIKQLDQEIRILGQLH-HPNIVEYYGSEVVGD--RLCIYMEYVHPGSLQKFM 1299
Query: 181 RKHRRI-PEPILSSMFQKLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAG 239
+ H + E ++ + + +L GL+YLH + +HRDIK ANLLV+ G K+ DFG+S
Sbjct: 1300 QDHCGVMTESVVRNFTRHILSGLAYLHSTK-TIHRDIKGANLLVDASGIVKLADFGVSKI 1476
Query: 240 L-ESSVAMCATFVGTVTYMSPERIR-------NESYSYPADIWSLGLALLECGTGEFPYT 291
L E S + + G+ +M+PE + N + + DIWSLG ++E TG+ P++
Sbjct: 1477 LTEKSYEL--SLKGSPYWMAPELMMAAMKNETNPTVAMAVDIWSLGCTIIEMLTGKPPWS 1650
Query: 292 ANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYE 351
G M ++L P +T SPE F++ C Q++P RP+A LL HPF+
Sbjct: 1651 EFPGH-QAMFKVLHRSPDIP--KTLSPEGQDFLEQCFQRNPADRPSAAVLLTHPFVQNLH 1821
Query: 352 TVEV 355
+V
Sbjct: 1822 EQDV 1833
>TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein kinase
{Arabidopsis thaliana}, partial (44%)
Length = 921
Score = 110 bits (274), Expect = 2e-24
Identities = 69/218 (31%), Positives = 112/218 (50%), Gaps = 9/218 (4%)
Frame = +2
Query: 146 YQGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRKH--RRIPEPILSSMFQKLLHGLS 203
+ L+ H +F S + + + +M GGS I++ EP+++++ +++L L
Sbjct: 59 HPNLLRAHCSFTAGHS--LWVVMPFMSGGSCLHIMKSSFPEGFDEPVIATVLREVLKALV 232
Query: 204 YLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESS---VAMCATFVGTVTYMSPE 260
YLH H +HRD+K N+L++ G K+ DFG+SA + + TFVGT +M+PE
Sbjct: 233 YLHAHGH-IHRDVKAGNILLDANGSVKMADFGVSACMFDTGDRQRSRNTFVGTPCWMAPE 409
Query: 261 RIRN-ESYSYPADIWSLGLALLECGTGEFPYTANEGPVNLMLQILDDPSPS---PSKQTF 316
++ Y + ADIWS G+ LE G P+ + P+ ++L L + P + F
Sbjct: 410 VMQQLHGYDFKADIWSFGITALELAHGHAPF-SKYPPMKVLLMTLQNAPPGLDYERDKRF 586
Query: 317 SPEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYETVE 354
S F + CL KDP RP++E+LL H F E
Sbjct: 587 SKSFKELVATCLVKDPKKRPSSEKLLKHHFFKHARATE 700
>TC88048 similar to GP|20302596|dbj|BAB91125. Ser/Thr kinase {Arabidopsis
thaliana}, partial (61%)
Length = 2349
Score = 107 bits (266), Expect = 1e-23
Identities = 80/267 (29%), Positives = 137/267 (50%), Gaps = 6/267 (2%)
Frame = +1
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKINIFE----KEKRQQLLTEIRTLCEAPCYQGLVEF 152
+G GAS V RA + +A ++ +++ E ++L EI L + ++ +++F
Sbjct: 100 LGKGASKTVYRAFDEYQGIEVAWNQVKLYDFLQSPEDLERLYCEIHLL-KTLKHKNIMKF 276
Query: 153 HGAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRH-L 211
+ ++ + I+ E G+L KH+R+ + ++L GL YLH +
Sbjct: 277 YTSWVDTANRNINFVTEMFTSGTLRQYRLKHKRVNIRAVKHWCIQILRGLLYLHSHDPPV 456
Query: 212 VHRDIKPANLLVN-LKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNESYSYP 270
+HRD+K N+ +N +GE KI D G++A L S A A VGT +M+PE + ESY+
Sbjct: 457 IHRDLKCDNIFINGNQGEVKIGDLGLAAILRKSHA--AHCVGTPEFMAPE-VYEESYNEL 627
Query: 271 ADIWSLGLALLECGTGEFPYTANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQK 330
DI+S G+ +LE T E+PY+ P + +++ P + PE F+D CL
Sbjct: 628 VDIYSFGMCILEMVTFEYPYSECTHPAQIYKKVISGKKPDALYKVKDPEVRQFVDKCL-A 804
Query: 331 DPDVRPTAEQLLFHPFITKYETVEVDL 357
+R +A++LL PF+ + + E DL
Sbjct: 805 TVSLRLSAKELLDDPFL-QIDDYEYDL 882
>TC79149 similar to PIR|T46059|T46059 MAP kinase - Arabidopsis thaliana,
partial (63%)
Length = 1671
Score = 105 bits (263), Expect = 3e-23
Identities = 81/289 (28%), Positives = 145/289 (50%), Gaps = 6/289 (2%)
Frame = +3
Query: 70 SVDESHDSDHSEKTYRCGSREMRIFGAIGSGASSVVQRAIHIPKHRIIALKKINIFE--- 126
S++E + + +T G R R +G GA V +AI +A ++ + E
Sbjct: 360 SMEEFKEENRYVETDPTG-RYGRFGDVLGKGAMKTVYKAIDEVLGIEVAWNQVRLNEVLN 536
Query: 127 -KEKRQQLLTEIRTLCEAPCYQGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRKHRR 185
+ Q+L +E+ L ++ ++ F+ ++ D+ + E GSL + RK++R
Sbjct: 537 TPDDLQRLYSEVHLLSTLK-HRSIMRFYTSWIDIDNKNFNFVTEMFTSGSLREYRRKYKR 713
Query: 186 IPEPILSSMFQKLLHGLSYLHG-VRHLVHRDIKPANLLVN-LKGEPKITDFGISAGLESS 243
+ + S +++L GL YLHG ++HRD K N+ VN G+ KI D G++A L+ S
Sbjct: 714 VSLQAIKSWARQILQGLVYLHGHDPPVIHRDPKCDNIFVNGHLGQVKIGDLGLAAILQGS 893
Query: 244 VAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLECGTGEFPYTANEGPVNLMLQI 303
+ + +GT +M+PE + E Y+ AD++S G+ +LE T ++PY+ P + ++
Sbjct: 894 QS-AHSVIGTPEFMAPE-MYEEEYNELADVYSFGMCVLEMLTSDYPYSECTNPAQIYKKV 1067
Query: 304 LDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYET 352
P + E FI CL+ + RP+A+ LL PF++ +T
Sbjct: 1068TSGKLPMSFFRIEDGEARRFIGKCLEPAAN-RPSAKDLLLEPFLSTDDT 1211
>TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Brassica
napus}, partial (34%)
Length = 851
Score = 98.6 bits (244), Expect(2) = 7e-23
Identities = 59/169 (34%), Positives = 99/169 (57%), Gaps = 1/169 (0%)
Frame = +3
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKINIFEKEKR-QQLLTEIRTLCEAPCYQGLVEFHGA 155
IG G+ V + ++ +A+K I++ E E + EI L + C + E++G+
Sbjct: 207 IGQGSFGDVYKGFDKELNKEVAIKVIDLEESEDDIDDIQKEISVLSQCRCPY-ITEYYGS 383
Query: 156 FYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHLVHRD 215
F + ++ I +EYM GGS+AD+L+ + E ++ + + LLH + YLH +HRD
Sbjct: 384 FL--NQTKLWIIMEYMAGGSVADLLQSGPPLDEMSIAYILRDLLHAVDYLHN-EGKIHRD 554
Query: 216 IKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRN 264
IK AN+L++ G+ K+ DFG+SA L +++ TFVGT +M+PE I+N
Sbjct: 555 IKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQN 701
Score = 26.9 bits (58), Expect(2) = 7e-23
Identities = 10/23 (43%), Positives = 15/23 (64%)
Frame = +1
Query: 264 NESYSYPADIWSLGLALLECGTG 286
++ Y+ ADIWSLG+ +E G
Sbjct: 703 SKGYNEKADIWSLGITAIEMAKG 771
>TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (45%)
Length = 976
Score = 103 bits (258), Expect = 1e-22
Identities = 60/189 (31%), Positives = 104/189 (54%), Gaps = 2/189 (1%)
Frame = +1
Query: 164 ISIALEYMDGGSLADILRKHRRI--PEPILSSMFQKLLHGLSYLHGVRHLVHRDIKPANL 221
+ I + Y +GG +A+ ++K + PE L +LL L YLH V H++HRD+K +N+
Sbjct: 304 VCIIIGYCEGGDMAETVKKANGVNFPEEKLCKWLVQLLMALDYLH-VNHILHRDVKCSNI 480
Query: 222 LVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALL 281
+ + ++ DFG+ A L +S + ++ VGT +YM PE + + Y +DIWSLG +
Sbjct: 481 FLTKNQDIRLGDFGL-AKLLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCVY 657
Query: 282 ECGTGEFPYTANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQL 341
E + A + + ++ ++ SP +S F + + L+K+P++RPTA +L
Sbjct: 658 EMAAHRPAFKAFD--IQALIHKINKSIVSPLPTMYSSAFRGLVKSMLRKNPELRPTAGEL 831
Query: 342 LFHPFITKY 350
L HP + Y
Sbjct: 832 LNHPHLQPY 858
>TC79461 similar to GP|9758254|dbj|BAB08753.1 contains similarity to
NRK-related kinase~gene_id:MWC10.1 {Arabidopsis
thaliana}, partial (88%)
Length = 1230
Score = 96.3 bits (238), Expect = 3e-20
Identities = 70/256 (27%), Positives = 127/256 (49%), Gaps = 6/256 (2%)
Frame = +1
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKINIF----EKEKRQQLLTEIRTLCEAPCYQGLVEF 152
+G GA V RA + +A ++ + E ++L +E+R L + + ++E
Sbjct: 142 LGCGAVKKVYRAFDQEEGIEVAWNQVKLRNFCDEPAMVERLYSEVRLL-RSLTNKNIIEL 318
Query: 153 HGAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRH-L 211
+ + + ++ E G+L + +KHR + L +++L GL+YLH +
Sbjct: 319 YSVWSDDRNNTLNFITEVCTSGNLREYRKKHRHVSMKALKKWSRQILKGLNYLHTHEPCI 498
Query: 212 VHRDIKPANLLVNLK-GEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNESYSYP 270
+HRD+ +N+ VN G+ KI D G++A + + T +GT +M+PE + +E Y+
Sbjct: 499 IHRDLNCSNVFVNGNVGQVKIGDLGLAA-IVGKNHIAHTILGTPEFMAPE-LYDEDYTEL 672
Query: 271 ADIWSLGLALLECGTGEFPYTANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQK 330
DI+S G+ +LE T E PY+ + + ++ P+ + E FI+ CL +
Sbjct: 673 VDIYSFGMCVLEMVTLEIPYSECDNVAKIYKKVSSGIRPAAMNKVKDSEVKEFIERCLAQ 852
Query: 331 DPDVRPTAEQLLFHPF 346
P RP+A +LL PF
Sbjct: 853 -PRARPSAAELLKDPF 897
>TC78081 similar to GP|16904226|gb|AAL30820.1 calcium/calmodulin-dependent
protein kinase CaMK3 {Nicotiana tabacum}, partial (58%)
Length = 1066
Score = 95.9 bits (237), Expect = 3e-20
Identities = 75/230 (32%), Positives = 116/230 (49%), Gaps = 8/230 (3%)
Frame = +2
Query: 136 EIRTLCEAPCYQGLVEFHGAFYTPDSGQISIALEYMDGGSLAD-ILRKHRRIPEPILSSM 194
E++ L + LV+F+ AF D + I +E +GG L D IL + + E ++
Sbjct: 83 EVKILRALNGHSNLVKFYDAF--EDQENVYIVMELCEGGELLDMILSRGGKYSEDDAKAV 256
Query: 195 FQKLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEP---KITDFGISAGLESSVAMCATFV 251
++L+ +++ H ++ +VHRD+KP N L K E K DFG+S + + V
Sbjct: 257 MVQILNVVAFCH-LQGVVHRDLKPENFLYTTKDESSELKAIDFGLSDFVRPDERL-NDIV 430
Query: 252 GTVTYMSPERIRNESYSYPADIWSLGLA--LLECGTGEFPYTANEGPVNLMLQILD--DP 307
G+ Y++PE + + SYS AD+WS+G+ +L CG+ F G +L+ D
Sbjct: 431 GSAYYVAPE-VLHRSYSTEADVWSIGVIAYILLCGSRPFWARTESGIFRAVLKADPGFDE 607
Query: 308 SPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYETVEVDL 357
P PS S E F+ L KDP R +A Q L HP+I Y V+V L
Sbjct: 608 GPWPS---LSSEAKDFVKRLLNKDPRKRISAAQALSHPWIRNYNDVKVPL 748
>TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine/threonine
protein kinase {Glycine max}, partial (90%)
Length = 2194
Score = 93.2 bits (230), Expect = 2e-19
Identities = 80/264 (30%), Positives = 124/264 (46%), Gaps = 6/264 (2%)
Frame = +3
Query: 89 REMRIFGAIGSGASSVVQRAIHIPKHRIIALKKIN---IFEKEKRQQLLTEIRTLCEAPC 145
R ++ +G G+ V+ A H+ +A+K +N I E +++ EI+ L
Sbjct: 108 RNYKMGKTLGIGSFGKVKIAEHVLTGHKVAIKILNRRKIKNMEMEEKVRREIKIL-RLFM 284
Query: 146 YQGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYL 205
+ ++ + TP I + +EY+ G L D + + R+ E S FQ+++ G+ Y
Sbjct: 285 HHHIIRLYEVVETPTD--IYVVMEYVKSGELFDYIVEKGRLQEDEARSFFQQIISGVEYC 458
Query: 206 HGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNE 265
H +VHRD+KP NLL++ K KI DFG+S + T G+ Y +PE I +
Sbjct: 459 HR-NMVVHRDLKPENLLLDSKWSVKIADFGLS-NIMRDGHFLKTSCGSPNYAAPEVISGK 632
Query: 266 SYSYP-ADIWSLGLAL--LECGTGEFPYTANEGPVNLMLQILDDPSPSPSKQTFSPEFCS 322
Y+ P D+WS G+ L L CGT F +E NL +I PS SP
Sbjct: 633 LYAGPEVDVWSCGVILYALLCGTLPFD---DENIPNLFKKIKGGIYTLPSH--LSPGARD 797
Query: 323 FIDACLQKDPDVRPTAEQLLFHPF 346
I L DP R T ++ HP+
Sbjct: 798 LIPRLLVVDPMKRMTIPEIRQHPW 869
>TC91968 similar to PIR|T02550|T02550 NPK1-related protein kinase homolog
T26B15.7 - Arabidopsis thaliana, partial (20%)
Length = 662
Score = 93.2 bits (230), Expect = 2e-19
Identities = 66/213 (30%), Positives = 113/213 (52%), Gaps = 6/213 (2%)
Frame = +2
Query: 97 IGSGASSVVQRAIHIPKH-RIIALKKINIFEKE---KRQQLLTEIRTLCEAPCYQGLVEF 152
IG G+++ V A + A+K + E K QQ+LT+++ C Q +V +
Sbjct: 17 IGRGSTATVYVADRCHSGGEVFAVKSTELHRSELLKKEQQILTKLK------CPQ-IVSY 175
Query: 153 HGAFYTPDSGQ--ISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRH 210
G T ++G ++ +EY G+L+D +R + E ++ +++L GL+YLH +
Sbjct: 176 QGCEVTFENGVQLFNLFMEYAPKGTLSDAVRNGNGMEEAMVGFYARQILLGLNYLH-LNG 352
Query: 211 LVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNESYSYP 270
+VH D+K N+LV +G KI+DFG + +E + + GT +M+PE R E Y
Sbjct: 353 IVHCDLKGQNVLVTEQGA-KISDFGCARRVEEELVIS----GTPAFMAPEVARGEEQGYA 517
Query: 271 ADIWSLGLALLECGTGEFPYTANEGPVNLMLQI 303
+D+W+LG LLE TG+ P+ P ++ +I
Sbjct: 518 SDVWALGCTLLEMITGKMPWQGFSDPAAVLYRI 616
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,038,354
Number of Sequences: 36976
Number of extensions: 256474
Number of successful extensions: 1955
Number of sequences better than 10.0: 500
Number of HSP's better than 10.0 without gapping: 1661
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1713
length of query: 526
length of database: 9,014,727
effective HSP length: 101
effective length of query: 425
effective length of database: 5,280,151
effective search space: 2244064175
effective search space used: 2244064175
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0263.11


