

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
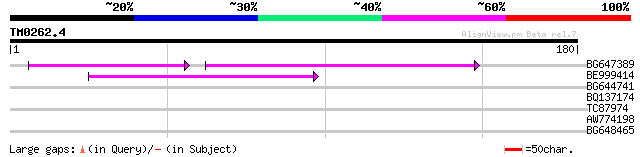
Query= TM0262.4
(180 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG647389 55 6e-13
BE999414 46 7e-06
BG644741 32 0.14
BQ137174 28 2.0
TC87974 homologue to PIR|S71750|S71750 import intermediate-assoc... 27 3.4
AW774198 26 7.5
BG648465 similar to GP|9955545|emb| ankyrin-repeat containing pr... 26 7.5
>BG647389
Length = 718
Score = 55.5 bits (132), Expect(2) = 6e-13
Identities = 30/87 (34%), Positives = 52/87 (59%)
Frame = +1
Query: 63 MFKGTAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYV 122
+ K W+ LP SI +++ + K + QFSASK +V+ +L+NVRQ E+L +Y+
Sbjct: 445 ILKEATQQWYMNLPRFSITGYQNMTHKLVHQFSASKHCKVSTTNLFNVRQDPNESLPEYL 624
Query: 123 KRYSAASVKIEELEP*ACARAFKDGLL 149
++ A++K+ + A AF++GLL
Sbjct: 625 V*FNNATIKVVNPKQELFAGAFQNGLL 705
Score = 34.3 bits (77), Expect(2) = 6e-13
Identities = 14/51 (27%), Positives = 29/51 (56%)
Frame = +2
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKC 57
P S + + +N+KSL+L ++ +DP +H F+ +M + A +++C
Sbjct: 281 PLSMDIWNAPVLENLKSLSLLSFDDKSDPVEHATTFNTQMAVIGALKSLRC 433
>BE999414
Length = 613
Score = 46.2 bits (108), Expect = 7e-06
Identities = 21/73 (28%), Positives = 37/73 (49%)
Frame = +3
Query: 26 LEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGTAMAWFTTLPLGSIVKFRD 85
+++Y G DP +H+ + + + AVKC++F + + AM WF L SI + D
Sbjct: 9 MDSYDGT*DPDEHMENIEVVLTYRSVRGAVKCKLFVTTLRRGAMTWFKNLRRNSIGSWGD 188
Query: 86 FSSKFLIQFSASK 98
+F F+ S+
Sbjct: 189LCHEFTTHFTVSR 227
>BG644741
Length = 735
Score = 32.0 bits (71), Expect = 0.14
Identities = 20/70 (28%), Positives = 30/70 (42%)
Frame = -2
Query: 58 RMFPSMFKGTAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERET 117
R+FP G A WFT LP SI + FL ++ K D + N L E+
Sbjct: 584 RVFPLSLMGEADIWFTELPYNSIFTWNQLRDVFLARYYPVSKKLNHNDRVNNFVALPGES 405
Query: 118 LKQYVKRYSA 127
+ R+++
Sbjct: 404 VSSSWDRFTS 375
>BQ137174
Length = 977
Score = 28.1 bits (61), Expect = 2.0
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 4/33 (12%)
Frame = -3
Query: 64 FKGT----AMAWFTTLPLGSIVKFRDFSSKFLI 92
+KGT +++WF TL G +V D +KFL+
Sbjct: 240 YKGTESIESLSWFKTLGEGVVVSIHDAQNKFLL 142
>TC87974 homologue to PIR|S71750|S71750 import intermediate-associated 100K
protein precursor - garden pea, partial (68%)
Length = 2331
Score = 27.3 bits (59), Expect = 3.4
Identities = 15/35 (42%), Positives = 19/35 (53%)
Frame = +1
Query: 13 REVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMV 47
REV I MK LA+EAY G P + + + S V
Sbjct: 2062 REVCILVIMKCLAVEAYWRGQSPHNSVFFVSFHFV 2166
>AW774198
Length = 554
Score = 26.2 bits (56), Expect = 7.5
Identities = 16/55 (29%), Positives = 26/55 (47%)
Frame = +2
Query: 9 SAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSM 63
SAV++ + N+KSL ++ Y G + P M+ S+ V C PS+
Sbjct: 257 SAVLKHLQPSTNLKSLTIKGYGGTSFPNWLGDNVFGNMLYLRISNCVNCLWLPSL 421
>BG648465 similar to GP|9955545|emb| ankyrin-repeat containing protein
{Arabidopsis thaliana}, partial (9%)
Length = 787
Score = 26.2 bits (56), Expect = 7.5
Identities = 17/63 (26%), Positives = 28/63 (43%)
Frame = +3
Query: 22 KSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGTAMAWFTTLPLGSIV 81
KSL + GT K+ L+F ++ SA S ++C P + T P S+
Sbjct: 264 KSLVVRCAKRGTTKKNEFLFFFNAVMASAKSVFLECSHLPPTLISHVLVAATCPP--SVT 437
Query: 82 KFR 84
+F+
Sbjct: 438 QFK 446
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.135 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,979,966
Number of Sequences: 36976
Number of extensions: 40841
Number of successful extensions: 236
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 235
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 236
length of query: 180
length of database: 9,014,727
effective HSP length: 90
effective length of query: 90
effective length of database: 5,686,887
effective search space: 511819830
effective search space used: 511819830
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0262.4


