

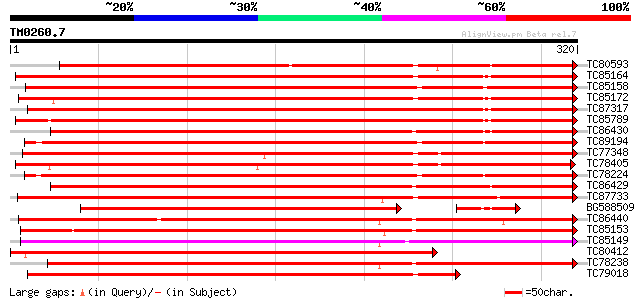
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0260.7
(320 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80593 similar to GP|1633130|pdb|1SCH|A Chain A Peanut Peroxid... 410 e-115
TC85164 similar to SP|P22195|PER1_ARAHY Cationic peroxidase 1 pr... 389 e-109
TC85158 similar to SP|P22195|PER1_ARAHY Cationic peroxidase 1 pr... 382 e-106
TC85172 weakly similar to GP|4760704|dbj|BAA77389.1 peroxidase 3... 369 e-103
TC87317 similar to GP|5381253|dbj|BAA82306.1 peroxidase {Nicotia... 345 1e-95
TC85789 similar to GP|5002344|gb|AAD37428.1| peroxidase 3 precur... 342 1e-94
TC86430 similar to GP|23315187|gb|AAN20368.1 Sequence 378 from p... 325 1e-89
TC89194 similar to GP|5453379|gb|AAD43561.1| bacterial-induced p... 323 4e-89
TC77348 similar to GP|571484|gb|AAB48986.1|| peroxidase precurso... 323 7e-89
TC78405 peroxidase precursor 319 8e-88
TC78224 similar to GP|5453379|gb|AAD43561.1| bacterial-induced p... 318 2e-87
TC86429 similar to GP|23315187|gb|AAN20368.1 Sequence 378 from p... 318 2e-87
TC87733 similar to GP|10177520|dbj|BAB10915. peroxidase {Arabido... 285 2e-77
BG588509 similar to GP|1633130|pdb Chain A Peanut Peroxidase, p... 268 2e-75
TC86440 weakly similar to PIR|S14268|S14268 peroxidase (EC 1.11.... 276 8e-75
TC85153 similar to GP|2245683|gb|AAC98519.1| peroxidase precurso... 268 3e-72
TC85149 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B... 268 3e-72
TC80412 similar to GP|5381253|dbj|BAA82306.1 peroxidase {Nicotia... 267 5e-72
TC78238 similar to PIR|T05723|T05723 peroxidase (EC 1.11.1.7) pr... 266 1e-71
TC79018 weakly similar to GP|10176746|dbj|BAB09977. peroxidase {... 265 2e-71
>TC80593 similar to GP|1633130|pdb|1SCH|A Chain A Peanut Peroxidase,
partial (68%)
Length = 1137
Score = 410 bits (1055), Expect = e-115
Identities = 205/294 (69%), Positives = 241/294 (81%), Gaps = 2/294 (0%)
Frame = +2
Query: 29 YNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSFTGEKN 88
Y +TCPKA+RTIR AV+ AV E RMGASLLRLHFHDCFVQGCDAS LLDDT +FTGEKN
Sbjct: 2 YGKTCPKAVRTIRKAVQDAVMNERRMGASLLRLHFHDCFVQGCDASALLDDTSNFTGEKN 181
Query: 89 SFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQLGRRDS 148
+FPNANSLRGFE+IDDIK QLE MCP VSC+DIL +AARD V LGGQRW++ LGRRDS
Sbjct: 182 AFPNANSLRGFELIDDIKSQLEDMCPNTVSCSDILALAARDGVAELGGQRWNVLLGRRDS 361
Query: 149 TTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRDRIYNE 208
TTA+L +N+ LPAPFL+L GLI+A+ KKGF A EMVTLSGAHTIGL RC F+R RIYNE
Sbjct: 362 TTANLSEANT-LPAPFLNLDGLITAFAKKGFTAEEMVTLSGAHTIGLVRCRFFRARIYNE 538
Query: 209 TNIDPSFAASMQVNCSFDGGDTDNNASPFDST--TQFKFDNAFYQNLLNQKGLVHSDQQL 266
TNIDP+FAA MQ C F+GG D+N SPFDS+ FDN +YQNL+ KGL+HSDQQL
Sbjct: 539 TNIDPAFAAKMQAECPFEGG--DDNFSPFDSSKPEAHDFDNGYYQNLVKSKGLIHSDQQL 712
Query: 267 YVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
+ NG+ +T++QV YS+N GRF +DFA+AM KMS+LSPLTG++G+IR NC VN
Sbjct: 713 FGNGT-STNAQVRRYSRNFGRFKKDFADAMFKMSMLSPLTGTEGEIRTNCHFVN 871
>TC85164 similar to SP|P22195|PER1_ARAHY Cationic peroxidase 1 precursor (EC
1.11.1.7). [Peanut] {Arachis hypogaea}, partial (92%)
Length = 1392
Score = 389 bits (1000), Expect = e-109
Identities = 199/317 (62%), Positives = 239/317 (74%)
Frame = +3
Query: 4 SLSCLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHF 63
+++ +I I +C VG SA LST FY+ TC L TI+ +++AV E RMGAS+LRLHF
Sbjct: 42 TMAKIIIPIILCFVGIVSAQLSTDFYSTTCSDVLSTIKREIDSAVGNEARMGASILRLHF 221
Query: 64 HDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADIL 123
HDCFVQGCDASVLLDDT SFTGEK + NANSLRGF+VID IK +LES+CP VSCADIL
Sbjct: 222 HDCFVQGCDASVLLDDTSSFTGEKTAGANANSLRGFDVIDTIKTELESLCPNTVSCADIL 401
Query: 124 TVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATE 183
+VAARDSV ALGG W +QLGRRDS TASL +NSDLP P DLSGLI+++ KGF E
Sbjct: 402 SVAARDSVVALGGPSWTVQLGRRDSITASLSLANSDLPGPGSDLSGLITSFDNKGFTPKE 581
Query: 184 MVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQF 243
MV LSG+HTIG + C F+R RIYN+ NID SFA S+Q NC GG D+N SP D+TT
Sbjct: 582 MVALSGSHTIGQASCRFFRTRIYNDDNIDSSFATSLQANCPTTGG--DDNLSPLDTTTPN 755
Query: 244 KFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLS 303
FDN+++QNL +QKGL SDQ L+ NG G+TDS V YS ++ F DFANAMVKM L+
Sbjct: 756 TFDNSYFQNLQSQKGLFSSDQALF-NG-GSTDSDVDEYSSDSSSFATDFANAMVKMGNLN 929
Query: 304 PLTGSDGQIRKNCRVVN 320
P+TGS+GQIR NCRV+N
Sbjct: 930 PITGSNGQIRTNCRVIN 980
>TC85158 similar to SP|P22195|PER1_ARAHY Cationic peroxidase 1 precursor (EC
1.11.1.7). [Peanut] {Arachis hypogaea}, partial (86%)
Length = 1161
Score = 382 bits (981), Expect = e-106
Identities = 198/311 (63%), Positives = 228/311 (72%)
Frame = +2
Query: 10 FLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQ 69
F++ CL+G SA LS+ FY TCP L TI+ V +A+ E RMGASLLRLHFHDCFVQ
Sbjct: 101 FIVLFCLIGTISAQLSSNFYFRTCPLVLSTIKKEVISALINERRMGASLLRLHFHDCFVQ 280
Query: 70 GCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARD 129
GCDASVLLDDT SF GEK + PNANSLRGF+VID IK ++E +CP VSCADIL VAARD
Sbjct: 281 GCDASVLLDDTSSFRGEKTAGPNANSLRGFDVIDKIKSEVEKLCPNTVSCADILAVAARD 460
Query: 130 SVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSG 189
SV ALGG W +QLGRRDSTTAS +NSDLP P DLSGLI+A+ KGF EMV LSG
Sbjct: 461 SVVALGGLSWTVQLGRRDSTTASFGLANSDLPGPGSDLSGLINAFNNKGFTPKEMVALSG 640
Query: 190 AHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAF 249
+HTIG + C F+R RIYNE NID SFA S+Q +C GGD N SP D+T+ FDNA+
Sbjct: 641 SHTIGEASCRFFRTRIYNENNIDSSFANSLQSSCPRTGGDL--NLSPLDTTSPNTFDNAY 814
Query: 250 YQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSD 309
++NL NQKGL HSDQ L+ TT SQV SY +N F DFANAM KM+ L PLTGS
Sbjct: 815 FKNLQNQKGLFHSDQVLF--DEVTTKSQVNSYVRNPLSFKVDFANAMFKMANLGPLTGSS 988
Query: 310 GQIRKNCRVVN 320
GQ+RKNCR VN
Sbjct: 989 GQVRKNCRSVN 1021
>TC85172 weakly similar to GP|4760704|dbj|BAA77389.1 peroxidase 3
{Scutellaria baicalensis}, partial (88%)
Length = 1282
Score = 369 bits (948), Expect = e-103
Identities = 194/336 (57%), Positives = 246/336 (72%), Gaps = 21/336 (6%)
Frame = +1
Query: 6 SCLIFLITICLVGKTSAV---------------------LSTIFYNETCPKALRTIRSAV 44
S +IF + ICL+G SA LS+ FY CP+AL IR +
Sbjct: 49 SSVIFCVMICLIGIVSASNVIPSATTLVSSLIPTPNYSELSSTFYGTRCPRALYIIRREI 228
Query: 45 EAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDD 104
AAVS++ R+GASLLRLHFHDCFVQGCDASVLL DT +F GE+N+ PNANSLRGFE ID
Sbjct: 229 IAAVSRDRRLGASLLRLHFHDCFVQGCDASVLLKDTPTFQGEQNARPNANSLRGFEFIDS 408
Query: 105 IKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPF 164
+K ++E++CP VVSCADIL VAARDSV LGG W ++LGRRDSTTA+ +A+NSDLP+PF
Sbjct: 409 LKAKIEAVCPNVVSCADILAVAARDSVATLGGPIWGVRLGRRDSTTANFNAANSDLPSPF 588
Query: 165 LDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCS 224
L+LSGLI+A+ KKGF+A EMV LSGAHTIG ++C +++RIYNE+NI+P + S+Q C
Sbjct: 589 LNLSGLIAAFKKKGFSADEMVALSGAHTIGKAKCAVFKNRIYNESNINPYYRRSLQNTCP 768
Query: 225 FDGGDTDNNASPFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKN 284
+GG DNN + DSTT FD+A+Y+NLL ++GL+HSDQ+LY NG G+TD +V +Y++N
Sbjct: 769 RNGG--DNNLANLDSTTPAFFDSAYYRNLLFKRGLLHSDQELY-NG-GSTDYKVLAYARN 936
Query: 285 AGRFLEDFANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
F DFA AM+KM LSPLTG+ GQIRK C VN
Sbjct: 937 PYLFRFDFAKAMIKMGNLSPLTGNQGQIRKYCSRVN 1044
>TC87317 similar to GP|5381253|dbj|BAA82306.1 peroxidase {Nicotiana
tabacum}, partial (94%)
Length = 1237
Score = 345 bits (886), Expect = 1e-95
Identities = 172/310 (55%), Positives = 217/310 (69%)
Frame = +3
Query: 11 LITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQG 70
L+ + +G +A LS +Y +CPK T++ V++A+SKE RMGASLLRL FHDCFV G
Sbjct: 114 LLVLVSIGSANANLSKDYYYSSCPKLFETVKCEVQSAISKETRMGASLLRLFFHDCFVNG 293
Query: 71 CDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDS 130
CD S+LLDDT SFTGEK + PN NS RGFEVID IK +E +CPG VSCADILT+ ARDS
Sbjct: 294 CDGSILLDDTSSFTGEKTANPNKNSARGFEVIDKIKSAVEKVCPGAVSCADILTITARDS 473
Query: 131 VFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGA 190
V LGG WD++LGRRD+ TAS A+N+D+PAP L+ LIS + G + ++V LSG
Sbjct: 474 VEILGGPTWDVKLGRRDARTASKSAANNDIPAPTSSLNQLISRFNALGLSTKDLVALSGG 653
Query: 191 HTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFY 250
HTIG +RC +R IYN++NID SFA + Q C G DNN +P D T FDN ++
Sbjct: 654 HTIGQARCTTFRAHIYNDSNIDTSFARTRQSGCPKTSGSGDNNLAPLDLATPTSFDNHYF 833
Query: 251 QNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDG 310
+NL++ KGL+HSDQQL+ NG G+TDS V YS F DF AM+KM +SPLTGS+G
Sbjct: 834 KNLVDSKGLLHSDQQLF-NG-GSTDSIVHEYSLYPSSFSSDFVTAMIKMGDISPLTGSNG 1007
Query: 311 QIRKNCRVVN 320
+IRK CR VN
Sbjct: 1008EIRKQCRSVN 1037
>TC85789 similar to GP|5002344|gb|AAD37428.1| peroxidase 3 precursor
{Phaseolus vulgaris}, partial (92%)
Length = 1467
Score = 342 bits (877), Expect = 1e-94
Identities = 180/319 (56%), Positives = 222/319 (69%), Gaps = 2/319 (0%)
Frame = +2
Query: 4 SLSCLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHF 63
+L C + + + G S LS FY + CP + + S V +AV++EPRMG SLLRLHF
Sbjct: 167 NLFCFVLFMFFLIDGSFSQ-LSENFYAKKCPNVFKAVNSVVHSAVAREPRMGGSLLRLHF 343
Query: 64 HDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADIL 123
HDCFV GCD SVLLDDT S GEK + PN +SLRGFEVID IK ++ES+CPGVVSCADI+
Sbjct: 344 HDCFVNGCDGSVLLDDTPSNKGEKTALPNKDSLRGFEVIDAIKSKVESVCPGVVSCADIV 523
Query: 124 TVAARDSVFALGGQRWDLQLGRRDSTTASL-DASNSDLPAPFLDLSGLISAYGKKGFNAT 182
+AARDSV LGG W ++LGRRDS TASL DA++ +P PF L+ LI+ + +G +
Sbjct: 524 AIAARDSVVNLGGPFWKVKLGRRDSKTASLNDANSGVIPPPFSTLNNLINRFKAQGLSTK 703
Query: 183 EMVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGD-TDNNASPFDSTT 241
+MV LSGAHTIG +RC YRDRIYN+TNID FA S Q NC G DNN + D T
Sbjct: 704 DMVALSGAHTIGKARCTVYRDRIYNDTNIDSLFAKSRQRNCPRKSGTIKDNNVAVLDFKT 883
Query: 242 QFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSL 301
FDN +Y+NL+N+KGL+HSDQ+L+ NG G+TDS V SYS N F DFA AM+KM
Sbjct: 884 PNHFDNLYYKNLINKKGLLHSDQELF-NG-GSTDSLVKSYSNNQNAFESDFAIAMIKMGN 1057
Query: 302 LSPLTGSDGQIRKNCRVVN 320
PLTGS+G+IRK CR N
Sbjct: 1058NKPLTGSNGEIRKQCRRAN 1114
>TC86430 similar to GP|23315187|gb|AAN20368.1 Sequence 378 from patent US
6410718, partial (93%)
Length = 1290
Score = 325 bits (833), Expect = 1e-89
Identities = 165/297 (55%), Positives = 209/297 (69%)
Frame = +2
Query: 24 LSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSF 83
LS FY +CP+ L + V A+ KE R+GASLLRLHFHDCFV GCDAS+LLDDT SF
Sbjct: 242 LSKDFYCSSCPELLSIVNQGVINAIKKETRIGASLLRLHFHDCFVNGCDASILLDDTSSF 421
Query: 84 TGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQL 143
GEK + N NS RGF VIDDIK +E CPGVVSCADILT+AARDSV LGG W++ L
Sbjct: 422 IGEKTAAANNNSARGFNVIDDIKANVEKACPGVVSCADILTLAARDSVVHLGGPSWNVGL 601
Query: 144 GRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRD 203
GRRDS TAS +N+ +PAPFL+LS L + + +G +A ++V LSGAHTIGL+RC+ +R
Sbjct: 602 GRRDSITASRSDANNSIPAPFLNLSALKTNFANQGLSAKDLVALSGAHTIGLARCVQFRA 781
Query: 204 RIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQNLLNQKGLVHSD 263
IYN++N+D F S+Q C G DN P D T FDN +++NLL +K L+HSD
Sbjct: 782 HIYNDSNVDSLFRKSLQNKCPRSG--NDNVLEPLDHQTPTHFDNLYFKNLLAKKALLHSD 955
Query: 264 QQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
Q+L+ NGS +TD+ V Y+ + +F + FA MVKMS + PLTGS+GQIR NCR +N
Sbjct: 956 QELF-NGS-STDNLVRKYATDNAKFFKAFAKGMVKMSSIKPLTGSNGQIRTNCRKIN 1120
>TC89194 similar to GP|5453379|gb|AAD43561.1| bacterial-induced peroxidase
precursor {Gossypium hirsutum}, partial (93%)
Length = 1186
Score = 323 bits (829), Expect = 4e-89
Identities = 166/312 (53%), Positives = 210/312 (67%)
Frame = +2
Query: 9 IFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFV 68
IF + C T+A L FY TCP +R + A++ E R+GAS+LRL FHDCFV
Sbjct: 113 IFSLLAC---STNAQLVNNFYGTTCPSLQTIVRREMTKAINNEARIGASILRLFFHDCFV 283
Query: 69 QGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAAR 128
GCD S+LLDDT +FTGEKN+ PN NS RGFEVID IK +E+ C VSCADIL +A R
Sbjct: 284 NGCDGSILLDDTSTFTGEKNAGPNKNSARGFEVIDAIKTSVEAACSATVSCADILALATR 463
Query: 129 DSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLS 188
D + LGG W + LGRRD+ TAS A+N+ +PAP DLS L + KG ++ LS
Sbjct: 464 DGIALLGGPSWIVPLGRRDARTASQSAANTQIPAPASDLSTLTKMFQNKGLTLRDLTVLS 643
Query: 189 GAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNA 248
GAHTIG + C F+R+RIYNETNID +FA + NC GGDT N +P DS + FDN
Sbjct: 644 GAHTIGQAECQFFRNRIYNETNIDTNFATLRKANCPLSGGDT--NLAPLDSVSPVTFDNN 817
Query: 249 FYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGS 308
+Y++L+ KGL++SDQ L+ NG G+ S V +YS N F DFA AMVKMS +SPLTG+
Sbjct: 818 YYRDLVANKGLLNSDQALF-NGVGSPVSLVRAYSINGFAFRRDFAFAMVKMSRISPLTGT 994
Query: 309 DGQIRKNCRVVN 320
+G+IRKNCR+VN
Sbjct: 995 NGEIRKNCRLVN 1030
>TC77348 similar to GP|571484|gb|AAB48986.1|| peroxidase precursor {Medicago
truncatula}, partial (95%)
Length = 1321
Score = 323 bits (827), Expect = 7e-89
Identities = 169/317 (53%), Positives = 221/317 (69%), Gaps = 4/317 (1%)
Frame = +1
Query: 8 LIFLITIC-LVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDC 66
++F++T ++ T A L+ +Y+ CPKAL I S V+ A+ +EPRMGASLLRLHFHDC
Sbjct: 70 VLFVVTFATILSPTIAKLTPNYYDRICPKALPIINSIVKQAIIREPRMGASLLRLHFHDC 249
Query: 67 FVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCP-GVVSCADILTV 125
FV GCD SVLLDDT +F GEK +FPN NS+RGFEV+D IK+ + C VVSCADIL +
Sbjct: 250 FVNGCDGSVLLDDTPTFIGEKTAFPNINSIRGFEVVDQIKEAVTKACKRDVVSCADILAI 429
Query: 126 AARDSVFALGGQRWDLQ--LGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATE 183
AARDSV LGG+++ Q LGRRDS AS DA+N++LP PF + S LI+ + G N +
Sbjct: 430 AARDSVAILGGKQYWYQVLLGRRDSRFASRDAANTNLPPPFFNFSQLITNFKSHGLNLKD 609
Query: 184 MVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQF 243
+V LSG HTIG S+C +RDRI+N+TNID +FAA++Q C GG D+N +PFDST
Sbjct: 610 LVVLSGGHTIGFSKCTNFRDRIFNDTNIDTNFAANLQKTCPKIGG--DDNLAPFDSTPN- 780
Query: 244 KFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLS 303
K D ++Y+ LL ++GL+HSDQ+L+ +D V YSKN+ F DF +M+KM L
Sbjct: 781 KVDTSYYKALLYKRGLLHSDQELFKGDGSQSDRLVQLYSKNSYAFAYDFGVSMIKMGNLK 960
Query: 304 PLTGSDGQIRKNCRVVN 320
PLTG G+IR NCR VN
Sbjct: 961 PLTGKKGEIRCNCRKVN 1011
>TC78405 peroxidase precursor
Length = 988
Score = 319 bits (818), Expect = 8e-88
Identities = 167/324 (51%), Positives = 223/324 (68%), Gaps = 8/324 (2%)
Frame = +2
Query: 4 SLSCLIFLITICLVGKTS-----AVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASL 58
S C IFL+ + + TS A+L+ FY+ CP+AL TI+S V A+ +E R+GASL
Sbjct: 20 SSPCQIFLVFVMVTLVTSLIPSNALLTPHFYDNVCPQALPTIKSVVLHAILREKRIGASL 199
Query: 59 LRLHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPG-VV 117
LRLHFHDCFV GCD SVLLDDT +FTGEK + PN NS+RGF V+D+IK ++ +C G VV
Sbjct: 200 LRLHFHDCFVNGCDGSVLLDDTPNFTGEKTALPNINSIRGFSVVDEIKAAVDKVCKGPVV 379
Query: 118 SCADILTVAARDSVFALGGQR--WDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYG 175
SCADIL AARDSV LGG + +++ LGRRD+ TAS A+N++LP+P + S LIS +
Sbjct: 380 SCADILATAARDSVAILGGPQFFYNVLLGRRDARTASKAAANANLPSPTFNFSQLISNFK 559
Query: 176 KKGFNATEMVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNAS 235
+G N ++V LSG HTIG +RC +R+RIYNETNIDP FAAS++ C +GG DNN +
Sbjct: 560 SQGLNVKDLVALSGGHTIGFARCTTFRNRIYNETNIDPIFAASLRKTCPRNGG--DNNLT 733
Query: 236 PFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANA 295
P D T + +N +Y++LL ++G++HSDQQL+ +D V YSKN F DF +
Sbjct: 734 PLDFTPT-RVENTYYRDLLYKRGVLHSDQQLFKGQGSESDKLVQLYSKNTFAFASDFKTS 910
Query: 296 MVKMSLLSPLTGSDGQIRKNCRVV 319
++KM + PLTG G+IR NCR V
Sbjct: 911 LIKMGNIKPLTGRQGEIRLNCRRV 982
>TC78224 similar to GP|5453379|gb|AAD43561.1| bacterial-induced peroxidase
precursor {Gossypium hirsutum}, partial (93%)
Length = 1276
Score = 318 bits (815), Expect = 2e-87
Identities = 164/312 (52%), Positives = 206/312 (65%)
Frame = +3
Query: 9 IFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFV 68
IF + C +A LS FY +TC +R+ + + KE RMGAS+LRL FHDCFV
Sbjct: 99 IFSLLAC--STINAQLSPNFYAKTCSNLQTIVRNEMIKVIQKEARMGASILRLFFHDCFV 272
Query: 69 QGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAAR 128
GCDAS+LLDD +F GEKNS PN S RGFEVID IK +E+ C VSCADIL +A R
Sbjct: 273 NGCDASILLDDKGTFVGEKNSGPNQGSARGFEVIDTIKTSVETACKATVSCADILALATR 452
Query: 129 DSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLS 188
D + LGG W + LGRRD+ TAS A+NS +P P DLS L + K ++ LS
Sbjct: 453 DGIALLGGPSWAVPLGRRDARTASQSAANSQIPGPSSDLSTLTRMFQNKSLTLNDLTVLS 632
Query: 189 GAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNA 248
GAHTIG + C F+R+RI+NE NID + A + NC GGDT N +PFDS T KFDN
Sbjct: 633 GAHTIGQTECQFFRNRIHNEANIDRNLATLRKRNCPTSGGDT--NLAPFDSVTPTKFDNN 806
Query: 249 FYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGS 308
+Y++L+ KGL+HSDQ L+ NG G+ S V YS++ F DFA AMVKMS +SPLTG+
Sbjct: 807 YYKDLIANKGLLHSDQVLF-NGGGSQISLVRKYSRDGAAFSRDFAAAMVKMSKISPLTGT 983
Query: 309 DGQIRKNCRVVN 320
+G+IRKNCR+VN
Sbjct: 984 NGEIRKNCRIVN 1019
>TC86429 similar to GP|23315187|gb|AAN20368.1 Sequence 378 from patent US
6410718, partial (90%)
Length = 1146
Score = 318 bits (814), Expect = 2e-87
Identities = 162/297 (54%), Positives = 203/297 (67%)
Frame = +1
Query: 24 LSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSF 83
L T FY +CP+ L + V A+ KE R+GASLLRLHFHDCFV GCDAS+LLDDT SF
Sbjct: 160 LCTDFYCNSCPELLSIVNQGVVNALKKETRIGASLLRLHFHDCFVNGCDASILLDDTSSF 339
Query: 84 TGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQL 143
GEK + N NS RGF VIDDIK +E CP VVSCADIL +AARDSV LGG WD+ L
Sbjct: 340 IGEKTAAANNNSARGFNVIDDIKASVEKACPKVVSCADILALAARDSVVHLGGPSWDVGL 519
Query: 144 GRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRD 203
GRRDS TAS +N+ +PAPF +LS L + + +G + ++V LSGAHTIGL+RC+ +R
Sbjct: 520 GRRDSITASRSDANNSIPAPFFNLSTLKTNFANQGLSVEDLVALSGAHTIGLARCVQFRA 699
Query: 204 RIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQNLLNQKGLVHSD 263
IYN++N+DP F S+Q C G DN PFD T FDN +++NLL +K L+HSD
Sbjct: 700 HIYNDSNVDPLFRKSLQNKCPRSG--NDNVLEPFDYQTPTHFDNLYFKNLLAKKTLLHSD 873
Query: 264 QQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
+L+ GS +T++ V Y+ N F + FA MVKMS + PLTGS+GQIR NCR N
Sbjct: 874 HELFNIGS-STNNLVRKYATNNAEFFKAFAEGMVKMSSIKPLTGSNGQIRINCRKTN 1041
>TC87733 similar to GP|10177520|dbj|BAB10915. peroxidase {Arabidopsis
thaliana}, partial (89%)
Length = 1270
Score = 285 bits (729), Expect = 2e-77
Identities = 152/323 (47%), Positives = 201/323 (62%), Gaps = 7/323 (2%)
Frame = +1
Query: 5 LSCLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFH 64
LS L F K + L FY+ +CP+A ++S + AV+KEPR+ ASLLRLHFH
Sbjct: 79 LSLLAFAPFCLCHKKMGSYLYPQFYDYSCPQAQNIVKSILANAVAKEPRIAASLLRLHFH 258
Query: 65 DCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILT 124
DCFV+GCDAS+LLD++ S EK S PN NS RGFEVID+IK LE CP VSCADIL
Sbjct: 259 DCFVKGCDASILLDNSGSIISEKGSNPNRNSARGFEVIDEIKYALEKECPHTVSCADILA 438
Query: 125 VAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEM 184
+AARDS GG W++ LGRRDS ASL SN+++PAP +++ + +G + ++
Sbjct: 439 IAARDSTVLAGGPNWEVPLGRRDSLGASLSGSNNNIPAPNNTFQTILTKFKLQGLDIVDL 618
Query: 185 VTLSGAHTIGLSRCIFYRDRIYNET-------NIDPSFAASMQVNCSFDGGDTDNNASPF 237
V LSG+HTIG SRC +R R+YN+T +D +AA ++ C GG D N
Sbjct: 619 VALSGSHTIGKSRCTSFRQRLYNQTGNGKQDFTLDQYYAAELRTQCPRSGG--DQNLFFL 792
Query: 238 DSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMV 297
D T KFDN +++NLL KGL+ SD+ L + + V Y++ F E FA +M+
Sbjct: 793 DYVTPTKFDNNYFKNLLAYKGLLSSDEILLTKNQESAE-LVKLYAERNDLFFEQFAKSMI 969
Query: 298 KMSLLSPLTGSDGQIRKNCRVVN 320
KM +SPLTGS G IR NCRV+N
Sbjct: 970 KMGNISPLTGSRGNIRTNCRVIN 1038
>BG588509 similar to GP|1633130|pdb Chain A Peanut Peroxidase, partial (68%)
Length = 741
Score = 268 bits (684), Expect(2) = 2e-75
Identities = 134/181 (74%), Positives = 150/181 (82%)
Frame = +3
Query: 41 RSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFE 100
+ AV+ AV E R+GASLLRLHF DCFVQGCD SVLLDDT SF GEKNS NANSLRGFE
Sbjct: 3 QQAVQNAVLGEARIGASLLRLHFQDCFVQGCDGSVLLDDTSSFKGEKNSLQNANSLRGFE 182
Query: 101 VIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDL 160
+IDDIK LE+MCP VVSCADILTVAARD+V LGGQ W++ LGRRDSTTASLDASNSD+
Sbjct: 183 LIDDIKSTLETMCPNVVSCADILTVAARDAVVLLGGQSWNVPLGRRDSTTASLDASNSDI 362
Query: 161 PAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQ 220
PAP L+L GLI+ + +K F A EMVTLSGAHTIG +RC +R RIYNETNIDPSFA S +
Sbjct: 363 PAPSLNLDGLIATFARKNFTALEMVTLSGAHTIGDARCTSFRGRIYNETNIDPSFAESKR 542
Query: 221 V 221
V
Sbjct: 543 V 545
Score = 32.3 bits (72), Expect(2) = 2e-75
Identities = 18/36 (50%), Positives = 28/36 (77%)
Frame = +2
Query: 253 LLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRF 288
L+++KGL+HSDQQL +NG +T QV +Y+ + G+F
Sbjct: 632 LVSKKGLLHSDQQL-LNGL-ST*HQVIAYTTDNGKF 733
>TC86440 weakly similar to PIR|S14268|S14268 peroxidase (EC 1.11.1.7)
neutral - horseradish, partial (87%)
Length = 1272
Score = 276 bits (706), Expect = 8e-75
Identities = 150/326 (46%), Positives = 203/326 (62%), Gaps = 11/326 (3%)
Frame = +2
Query: 6 SCLIF-LITICLVGKT-SAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHF 63
SC F LI + ++ T + L+T FY +CP + +R V A+ E RMGASLLRLHF
Sbjct: 104 SCYYFVLINMFMLHFTVRSQLTTDFYKSSCPNLTKIVRKEVVKALKNEMRMGASLLRLHF 283
Query: 64 HDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADIL 123
HDCFV GCD S+LLD D F EK++FPN NS+RGF+V+D IK +ES C GVVSCADIL
Sbjct: 284 HDCFVNGCDGSILLDGGDDF--EKSAFPNINSVRGFDVVDTIKTAVESACSGVVSCADIL 457
Query: 124 TVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATE 183
+AARDSV GG W + LGRRD T ++ +N LP+PF L ++S + G N T+
Sbjct: 458 AIAARDSVLLSGGPSWSVMLGRRDGTISNGSLANVVLPSPFDPLDTIVSKFTNVGLNLTD 637
Query: 184 MVTLSGAHTIGLSRCIFYRDRIYN-------ETNIDPSFAASMQVNCSFDGGDTDNNASP 236
+V+LSGAHTIG +RC + +R++N ++ ++ +Q C G N +
Sbjct: 638 VVSLSGAHTIGRARCALFSNRLFNFSGTGSPDSTLETGMLTDLQNLCPQTG--DGNTTAV 811
Query: 237 FDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQ--VTSYSKNAGRFLEDFAN 294
D + FDN +Y+NLLN KGL+ SDQ L + S+ V SY+ NA F DF
Sbjct: 812 LDRNSTDLFDNHYYKNLLNGKGLLSSDQILISTDEANSTSKPLVQSYNDNATLFFGDFVK 991
Query: 295 AMVKMSLLSPLTGSDGQIRKNCRVVN 320
+M+KM ++P TGSDG+IRK+CRV+N
Sbjct: 992 SMIKMGNINPKTGSDGEIRKSCRVIN 1069
>TC85153 similar to GP|2245683|gb|AAC98519.1| peroxidase precursor {Glycine
max}, complete
Length = 1413
Score = 268 bits (684), Expect = 3e-72
Identities = 143/322 (44%), Positives = 198/322 (61%), Gaps = 8/322 (2%)
Frame = +3
Query: 7 CLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSK-EPRMGASLLRLHFHD 65
C + ++ L ++A L FY +TCPK L +I V V+K +PRM AS++RLHFHD
Sbjct: 78 CCVVVVFGGLPFSSNAQLDPYFYGKTCPK-LHSIAFKVLRKVAKTDPRMPASIIRLHFHD 254
Query: 66 CFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTV 125
CFVQGCDASVLL++T + E+++FPN NSLRG +VI+ IK ++E CP VSCADILT+
Sbjct: 255 CFVQGCDASVLLNNTATIVSEQDAFPNINSLRGLDVINQIKTKVEKACPNRVSCADILTL 434
Query: 126 AARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMV 185
A+ S GG W++ LGRRDS TA+ +N +LP P L L SA+ +G N ++V
Sbjct: 435 ASGISSVLTGGPGWEVPLGRRDSLTANQSLANQNLPGPNFSLDRLKSAFAAQGLNTVDLV 614
Query: 186 TLSGAHTIGLSRCIFYRDRIYNETN-------IDPSFAASMQVNCSFDGGDTDNNASPFD 238
LSGAHT G +RC+F DR+YN N +D ++ ++ C +G T NN FD
Sbjct: 615 ALSGAHTFGRARCLFILDRLYNFNNTGKPDPTLDTTYLQQLRNQCPQNG--TGNNRVNFD 788
Query: 239 STTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVK 298
TT D FY NL +KGL+ SDQ+L+ T S V S++ + F ++F N+M+K
Sbjct: 789 PTTPDTLDKNFYNNLQGKKGLLQSDQELFSTPGADTISIVNSFANSQNVFFQNFINSMIK 968
Query: 299 MSLLSPLTGSDGQIRKNCRVVN 320
M + LTG G+IRK C +N
Sbjct: 969 MGNIDVLTGKKGEIRKQCNFIN 1034
>TC85149 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B precursor
- alfalfa, partial (95%)
Length = 1274
Score = 268 bits (684), Expect = 3e-72
Identities = 138/321 (42%), Positives = 188/321 (57%), Gaps = 7/321 (2%)
Frame = +2
Query: 7 CLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDC 66
C + ++ L + A L FY +TCPK IR + +PRM ASL+RLHFHDC
Sbjct: 53 CCVVVVLGGLPFSSDAQLDPSFYRDTCPKVHSIIREVIRNVSKTDPRMLASLVRLHFHDC 232
Query: 67 FVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVA 126
FV GCDASVLL+ TD+ E+ +FPN NSLRG +V++ IK +E CP VSCADIL ++
Sbjct: 233 FVLGCDASVLLNKTDTIVSEQEAFPNINSLRGLDVVNQIKTAVEKACPNTVSCADILALS 412
Query: 127 ARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVT 186
A+ S G W + LGRRD TA+ +N +LPAPF L L SA+ +G + T++V
Sbjct: 413 AQISSILADGPNWKVPLGRRDGLTANQSLANQNLPAPFNSLDQLKSAFAAQGLSTTDLVA 592
Query: 187 LSGAHTIGLSRCIFYRDRIYN-------ETNIDPSFAASMQVNCSFDGGDTDNNASPFDS 239
LSGAHT G +RC F DR+YN + ++ ++ ++ C G NN + FD
Sbjct: 593 LSGAHTFGRARCTFITDRLYNFSSTGKPDPTLNTTYLQELRKIC--PNGGPPNNLANFDP 766
Query: 240 TTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKM 299
TT KFD +Y NL +KGL+ SDQ+L+ T S V +S + F + F AM+KM
Sbjct: 767 TTPDKFDKNYYSNLQGKKGLLQSDQELFSTSGADTISIVNKFSADKNAFFDSFEAAMIKM 946
Query: 300 SLLSPLTGSDGQIRKNCRVVN 320
+ LTG G+IRK+C VN
Sbjct: 947 GNIGVLTGKKGEIRKHCNFVN 1009
>TC80412 similar to GP|5381253|dbj|BAA82306.1 peroxidase {Nicotiana
tabacum}, partial (71%)
Length = 817
Score = 267 bits (682), Expect = 5e-72
Identities = 131/246 (53%), Positives = 174/246 (70%), Gaps = 5/246 (2%)
Frame = +3
Query: 1 SRGSLSC-----LIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMG 55
S LSC ++ + ++G +A LST FY++TCPK ++ V++A+SKE R+G
Sbjct: 72 SMAPLSCSRITMFSLVLFVLIIGSVNAQLSTNFYSKTCPKLSSIVQRQVQSAISKEARIG 251
Query: 56 ASLLRLHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPG 115
AS+LRL FHDCFV GCD S+LLDDT +FTGEKN+ PN NS+RGF+VID+IK +E++CPG
Sbjct: 252 ASILRLFFHDCFVNGCDGSILLDDTSNFTGEKNALPNKNSVRGFDVIDNIKTAVENVCPG 431
Query: 116 VVSCADILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYG 175
VVSCADIL +AA DSV LGG W+++LGRRD+TTAS +N+ +P P +L+ L S +
Sbjct: 432 VVSCADILAIAATDSVAILGGPTWNVKLGRRDATTASQSDANTAIPRPTSNLNILTSMFK 611
Query: 176 KKGFNATEMVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNAS 235
G + ++V LSGAHTIG +RC +R RIYNETNID SFA++ Q NC G DNN +
Sbjct: 612 NVGLSTKDLVALSGAHTIGQARCTTFRVRIYNETNIDTSFASTRQSNCPKTSGSGDNNLA 791
Query: 236 PFDSTT 241
P D T
Sbjct: 792 PLDLHT 809
>TC78238 similar to PIR|T05723|T05723 peroxidase (EC 1.11.1.7) precursor
seed coat - soybean, partial (90%)
Length = 1243
Score = 266 bits (679), Expect = 1e-71
Identities = 135/306 (44%), Positives = 198/306 (64%), Gaps = 7/306 (2%)
Frame = +2
Query: 22 AVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTD 81
A LS FY++TCP + + A +PR+GASL+RLHFHDCFVQGCD SVLL++T+
Sbjct: 89 AQLSPSFYSQTCPFLYPIVFRVIFEASLTDPRIGASLIRLHFHDCFVQGCDGSVLLNNTN 268
Query: 82 SFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDL 141
+ E+++ PN NSLRG +V++ IK +E+ CP VSCADILT+AA+ + GG W +
Sbjct: 269 TIVSEQDALPNINSLRGLDVVNQIKTAVENECPATVSCADILTIAAQVASVLGGGPSWQI 448
Query: 142 QLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFY 201
LGRRDS TA+ +N +LPAPF L L +A+ +G N T++VTLSGAHT G ++C +
Sbjct: 449 PLGRRDSLTANQALANQNLPAPFFTLDQLKAAFLVQGLNTTDLVTLSGAHTFGRAKCSTF 628
Query: 202 RDRIYN-------ETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQNLL 254
+R+YN + ++ ++ +++ C +G T NN + D TT +FDN FY NL
Sbjct: 629 INRLYNFNSTGNPDQTLNTTYLQTLREICPQNG--TGNNLTNLDLTTPNQFDNKFYSNLQ 802
Query: 255 NQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRK 314
+ KGL+ SDQ+L+ + T + V S+S N F E+F +M+KM+ +S LTG++G+IR
Sbjct: 803 SHKGLLQSDQELFSTPNADTIAIVNSFSSNQALFFENFRVSMIKMANISVLTGNEGEIRL 982
Query: 315 NCRVVN 320
C +N
Sbjct: 983 QCNFIN 1000
>TC79018 weakly similar to GP|10176746|dbj|BAB09977. peroxidase {Arabidopsis
thaliana}, partial (66%)
Length = 831
Score = 265 bits (676), Expect = 2e-71
Identities = 136/246 (55%), Positives = 167/246 (67%), Gaps = 2/246 (0%)
Frame = +1
Query: 11 LITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQG 70
++ L KT + L+T +Y+ TCP AL TIRS VEAAV KE RMG SLLRLHFHDCFV G
Sbjct: 100 IVLAALATKTFSALTTNYYDYTCPNALSTIRSVVEAAVEKENRMGGSLLRLHFHDCFVNG 279
Query: 71 CDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMC-PGVVSCADILTVAARD 129
CD S+LLD T S EKN+ PN NS RGFEV+D+IK ++ C VVSCADIL +AARD
Sbjct: 280 CDGSILLDSTPSMDSEKNANPNINSARGFEVVDEIKDAVDKACGKPVVSCADILAIAARD 459
Query: 130 SVFALGGQRWDLQLGRRDSTTAS-LDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLS 188
SV ALGG W ++LGRRDS TAS DA + ++P P LS LI + +G N ++V LS
Sbjct: 460 SVVALGGPSWKVKLGRRDSKTASRADADSGNIPGPAFSLSQLIKNFDNQGLNEKDLVALS 639
Query: 189 GAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNA 248
GAHTIG SRC +RDRIY + NI+ FA +Q C +GG D+N +P DS T FD A
Sbjct: 640 GAHTIGFSRCFLFRDRIYKDNNINAYFAKQLQNVCPREGG--DSNLAPLDSVTSANFDVA 813
Query: 249 FYQNLL 254
+Y L+
Sbjct: 814 YYSQLI 831
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,372,101
Number of Sequences: 36976
Number of extensions: 117131
Number of successful extensions: 852
Number of sequences better than 10.0: 146
Number of HSP's better than 10.0 without gapping: 680
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 697
length of query: 320
length of database: 9,014,727
effective HSP length: 96
effective length of query: 224
effective length of database: 5,465,031
effective search space: 1224166944
effective search space used: 1224166944
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0260.7


