

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0260.2
(694 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
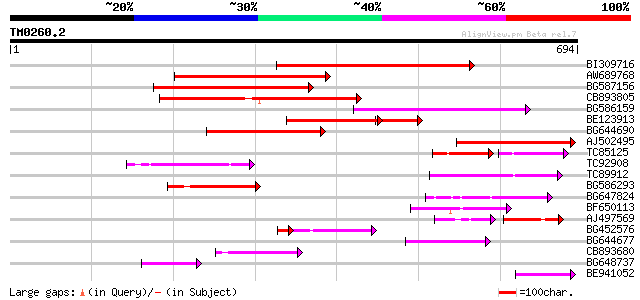
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 285 4e-77
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 200 2e-51
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 181 8e-46
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 177 9e-45
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 160 2e-39
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 113 4e-36
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 123 3e-28
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 118 9e-27
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 68 8e-25
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 105 8e-23
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 104 1e-22
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 97 2e-20
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 97 2e-20
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 82 7e-16
AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F... 54 2e-14
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 63 5e-12
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 65 9e-11
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 65 1e-10
BG648737 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberos... 63 4e-10
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 62 6e-10
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 285 bits (729), Expect = 4e-77
Identities = 140/242 (57%), Positives = 180/242 (73%)
Frame = +2
Query: 327 QVCLLQKSLYGLKQASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDV 386
+VC LQKS+YGLKQASRQWY+ L +L S GY Q D +L K +S T LL+YVDD+
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 387 LLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAG 446
+L GN EIQ VK FL +F+IKDLG L++FLGLEVARS +GI L QRKY LELL D+G
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSG 376
Query: 447 LLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLA 506
L K TP D +L L + L +D T YRRL+G+L+YLTTTRPD+++ VQQLSQ+++
Sbjct: 377 NLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVS 556
Query: 507 SPTDLHQQAATRVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFL 566
P +H QAA RVL+Y+K AP +GLF+SAT++LKL +++DSDWA CP TR+S+ GY +FL
Sbjct: 557 KPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFL 736
Query: 567 GS 568
GS
Sbjct: 737 GS 742
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 200 bits (508), Expect = 2e-51
Identities = 96/191 (50%), Positives = 128/191 (66%)
Frame = +1
Query: 202 WRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQ 261
W AM E +AL N TW +V LPP KK IGCKWVYRVK DGS++++KARLVAKG++Q
Sbjct: 97 WLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQ 276
Query: 262 VEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVS 321
G DY +TFSPV K T+RL+L +A + W + Q+D++NAFL+G L EE+YM P G
Sbjct: 277 TLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFE 456
Query: 322 SPRPNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLL 381
+ + VC L KSLYGLKQA R WY L +A G+ + D +LL+ + + L +
Sbjct: 457 AANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXI 636
Query: 382 YVDDVLLTGNS 392
YVDD+L+TG+S
Sbjct: 637 YVDDILITGSS 669
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 181 bits (459), Expect = 8e-46
Identities = 86/197 (43%), Positives = 128/197 (64%), Gaps = 1/197 (0%)
Frame = -1
Query: 177 HYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWV 236
H AF N++ P +Y +A + KEW++++ E A+ +N+TW +LP KK + +W+
Sbjct: 597 HCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWI 418
Query: 237 YRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQ 296
+ +K+K DGSI+R K RLVA+G+T G DY++TF+PVAK T+R++L+LA + GW L Q
Sbjct: 417 FTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQ 238
Query: 297 LDVDNAFLHGTLDEEIYMRLPPGVSS-PRPNQVCLLQKSLYGLKQASRQWYNTLVAALKS 355
+DV NAFL G L++E+YM PPG+ + V L+K++YGLKQ+ R WYN L L
Sbjct: 237 MDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNG 58
Query: 356 LGYEQCPHDHTLLLKRT 372
G+ + DHTL T
Sbjct: 57 RGFRKSELDHTLFTLTT 7
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 177 bits (450), Expect = 9e-45
Identities = 95/256 (37%), Positives = 154/256 (60%), Gaps = 9/256 (3%)
Frame = +3
Query: 184 VSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQ 243
++ T +PTT+ +A + ++WR +M+ E+EA RNNTW + DL K IG KW+++ K +
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 244 DGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAF 303
+G I++YKARLVAKGY+Q G+DY + F+PVA+ T+R+++ALAA Q+ D
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAA-------QIKRDGVC 362
Query: 304 L------HGTLDE--EIYMRLPPGVSSPRPNQVCLLQKSLYGLKQASRQWYNTLVAALKS 355
+ H ++ ++ + V R ++++LYGLKQA R WY+ + A
Sbjct: 363 IS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS-*RVKRALYGLKQAPRAWYSRIEAYFTK 539
Query: 356 LGYEQCPHDHTLLLKRT-STSITALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGD 414
G+E+CP++HTL +K + I + LYVDD++ GN + K + +F + DLG
Sbjct: 540 EGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGK 719
Query: 415 LKFFLGLEVARSAKGI 430
+ +FLG+EV ++ KGI
Sbjct: 720 MHYFLGVEVTQNEKGI 767
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 160 bits (405), Expect = 2e-39
Identities = 81/217 (37%), Positives = 126/217 (57%)
Frame = +1
Query: 421 LEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYRRL 480
+EV ++ +GI + QRKY +LL G+ R P+ L + + D T Y+++
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 481 VGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATASLK 540
VG L+YL TRPDL YV+ +S+++ PT+LH A RVLRY+ G G+ + S K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 541 LQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVCEV 600
L+AY+DSD+AG D R+S +GY L S +SW +KKQ V+ S+++AE+ A A C+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 601 QWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFH 637
W+ + + L + ++CDN S I +++NP H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 113 bits (283), Expect(2) = 4e-36
Identities = 54/118 (45%), Positives = 80/118 (67%), Gaps = 1/118 (0%)
Frame = +1
Query: 340 QASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLL-YVDDVLLTGNSFEEIQA 398
Q+ R W++ +K GY QC DH + +K +ST A+L+ YVDD+ LTG+ + I+
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 399 VKSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTP 456
+K+ L +F IKDLG+LK+FLG+EVAR KG ++QRKY L+LL + ++GCK +R P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 57.0 bits (136), Expect(2) = 4e-36
Identities = 27/58 (46%), Positives = 39/58 (66%)
Frame = +2
Query: 448 LGCKPVRTPMDSTLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYL 505
L KP TPMD+T+ L L D Y+RLVG+L+YL+ TRPD+++VV +SQ++
Sbjct: 329 LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 123 bits (308), Expect = 3e-28
Identities = 64/146 (43%), Positives = 93/146 (62%), Gaps = 1/146 (0%)
Frame = -2
Query: 242 KQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDN 301
KQ I R K++LV +GY Q EG+DY + FSPVA+ +R+L+A AA G+ L+Q+DV +
Sbjct: 439 KQA*VITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKS 260
Query: 302 AFLHGTLDEEIYMRLPPGVSSPR-PNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGYEQ 360
AF++G L EE++++ PPG PN V L K+LYGLKQA R WY L L G+++
Sbjct: 259 AFINGDLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKR 80
Query: 361 CPHDHTLLLKRTSTSITALLLYVDDV 386
D+TL L + + + +YVDD+
Sbjct: 79 GKIDNTLFLLKRE*ELLIIQVYVDDI 2
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 118 bits (295), Expect = 9e-27
Identities = 57/146 (39%), Positives = 88/146 (60%)
Frame = +2
Query: 547 SDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYL 606
SDWAG +TR+S +GY+ LG+ ISW +KKQ V+ S++EAEY A + + WL +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 607 FQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGLIHLLPVST 666
+ +H P ++CDN+SAI +++NP FH R+KHI++ H +R + + + T
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 667 RDQLADIFTKPLPPSLFPTFVLKLGL 692
+++ADIFTKPL F LG+
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGM 439
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 68.2 bits (165), Expect(2) = 8e-25
Identities = 33/85 (38%), Positives = 49/85 (56%)
Frame = +3
Query: 599 EVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGL 658
E W+ L + L + + ++CD+QSA+HIA+NP FH RTKHI + H VR ++ G
Sbjct: 249 EAIWMQRLMEELGHKQE-QITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVEEGS 425
Query: 659 IHLLPVSTRDQLADIFTKPLPPSLF 683
+ + + T D LAD TK + F
Sbjct: 426 VDMQKIHTNDNLADAMTKSINTDKF 500
Score = 64.3 bits (155), Expect(2) = 8e-25
Identities = 32/75 (42%), Positives = 46/75 (60%)
Frame = +1
Query: 518 RVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKK 577
R++RY+KG G + F + L ++ Y DSD+AG D R+S GY L +SW +K
Sbjct: 7 RIMRYIKGTSGVAVCFGGS-ELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSKL 183
Query: 578 QTTVSRSSSEAEYRA 592
QT V+ S++EAEY A
Sbjct: 184 QTVVALSTTEAEYMA 228
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 105 bits (261), Expect = 8e-23
Identities = 63/156 (40%), Positives = 91/156 (57%)
Frame = +2
Query: 144 SSIAALNSSSKVSSTGIKHPLSSVLSYTNLSPKHYAFTTNVSSTVEPTTYTQAAQCKEWR 203
S + LNSSSK + + + Y+ S KH A ++S +EP Y QAAQ +EW
Sbjct: 164 SHLVTLNSSSK------SYHIFHYVGYS-FSAKHRASLAAITSNIEPKNYVQAAQ*QEWL 322
Query: 204 DAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVE 263
AM++E++ L NNT + L KK + C+ VY++ HK +G I++YKA+LVAK + QVE
Sbjct: 323 AAMEQEIQVLEENNTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVE 502
Query: 264 GLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDV 299
G D+ D K R LL +AA++G LH +DV
Sbjct: 503 GEDF*D-LCLSNKDDNCRCLLTIAAAKG*QLHLMDV 607
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 104 bits (259), Expect = 1e-22
Identities = 57/165 (34%), Positives = 99/165 (59%), Gaps = 2/165 (1%)
Frame = +1
Query: 514 QAATRVLRYVKGAPGQGLFFSATASLK--LQAYSDSDWAGCPDTRRSINGYSIFLGSSLI 571
QA VL+Y+ + L ++ A + L+ Y D+D+AG DTR+S++G+ L + I
Sbjct: 1 QALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTI 180
Query: 572 SWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIA 631
SW+ +Q+ V+ S+++AEY A V + WL + L + + V + CD+QSAIH+A
Sbjct: 181 SWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEY-VKIHCDSQSAIHLA 357
Query: 632 QNPTFHERTKHIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTK 676
+ +HERTKHI++ H +R +++ I + +++ + AD+FTK
Sbjct: 358 NHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 97.4 bits (241), Expect = 2e-20
Identities = 48/113 (42%), Positives = 73/113 (64%)
Frame = +2
Query: 194 TQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKAR 253
T +C E RD + + + L +V P KPIG +W+Y++K +DG++ +YKAR
Sbjct: 2 TSLDRCNEGRDRIYYQKQTLK------LVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKAR 163
Query: 254 LVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAFLHG 306
LVAKGY + +G+D+ + F+PV + T+ LLLALAA+ G +H +DV AFL+G
Sbjct: 164 LVAKGYVKQQGIDFDEVFAPVVRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 97.1 bits (240), Expect = 2e-20
Identities = 62/157 (39%), Positives = 88/157 (55%), Gaps = 1/157 (0%)
Frame = -3
Query: 509 TDLHQQAATRVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGS 568
T H QAA ++L Y+K +P Q +FF + +K A+ DSD ++ S+ S
Sbjct: 599 TAQHPQAAIQIL-YLKISPSQ*IFFPS*IQIK--AFCDSD*ID--QAA*TLENQSVIFAS 435
Query: 569 SLISWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAI 628
S + R + + YR++ + +CE++WL+YL L P L+CDNQSA
Sbjct: 434 S*ATHSYAGNLKKKRYNFKI-YRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCDNQSAA 258
Query: 629 -HIAQNPTFHERTKHIELDCHVVRTKLQAGLIHLLPV 664
HIA N +F ERTKHIELDCH+VR KLQ L H+L +
Sbjct: 257 RHIAANSSFLERTKHIELDCHIVRVKLQLKLFHILHI 147
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 82.0 bits (201), Expect = 7e-16
Identities = 51/127 (40%), Positives = 65/127 (51%), Gaps = 3/127 (2%)
Frame = +1
Query: 491 RPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATAS---LKLQAYSDS 547
RPD+ Y V +S+++ P H AA R+LRYV+G GL F A +L YSDS
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 548 DWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYLF 607
DW G RRS +GY + ISW TKKQ + SS EAEY A + WL +
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 608 QFLHLRV 614
+ L V
Sbjct: 472 KELKCEV 492
>AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F21P8.50 -
Arabidopsis thaliana, partial (4%)
Length = 723
Score = 53.9 bits (128), Expect(2) = 2e-14
Identities = 29/74 (39%), Positives = 45/74 (60%)
Frame = +2
Query: 605 YLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGLIHLLPV 664
++F + + V +CDN SA+HIA N FHERT H E D ++V+ + ++ L+P
Sbjct: 242 FIFYMISAKHST*VLQYCDNISALHIAANMVFHERT*HRETDPYIVQ---GSRMLQLMPS 412
Query: 665 STRDQLADIFTKPL 678
+++DQ A TKPL
Sbjct: 413 ASKDQPAYSLTKPL 454
Score = 43.1 bits (100), Expect(2) = 2e-14
Identities = 33/75 (44%), Positives = 42/75 (56%)
Frame = +3
Query: 520 LRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQT 579
L Y+K PG+ +F S ++ S CP + R + FL SSLISW++KKQ
Sbjct: 15 LHYLK-TPGKCIFVSNASNPHFNRGS------CPYSIR*TTEFC-FLSSSLISWKSKKQC 170
Query: 580 TVSRSSSEAEYRALA 594
VSRS SEA RALA
Sbjct: 171 VVSRSFSEA**RALA 215
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like
protein {Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 63.2 bits (152), Expect(2) = 5e-12
Identities = 40/106 (37%), Positives = 60/106 (55%)
Frame = +2
Query: 343 RQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSF 402
R+ Y L++ L SLGY+Q P+DH++ T L+Y+DD++ + E Q VKS
Sbjct: 236 RK*Y*KLLSTLISLGYKQSPNDHSIF--SFGRRFTIFLVYLDDIVFARDDHSETQLVKSH 409
Query: 403 LHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLL 448
L F I LG L ++G+++A S I Q KY +ELL ++G L
Sbjct: 410 LDKNFIIIGLGTL-HYVGVKIA*SES*IIDDQCKYTIELLEESGQL 544
Score = 26.2 bits (56), Expect(2) = 5e-12
Identities = 11/20 (55%), Positives = 13/20 (65%)
Frame = +3
Query: 328 VCLLQKSLYGLKQASRQWYN 347
VC S+YGLK A RQ Y+
Sbjct: 165 VCKFHNSIYGLKHAHRQ*YS 224
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 65.1 bits (157), Expect = 9e-11
Identities = 34/104 (32%), Positives = 57/104 (54%)
Frame = -3
Query: 485 LYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATASLKLQAY 544
+ LT P++ + + LS+Y ++PT H + +Y+KG GLF+S S L Y
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 545 SDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEA 588
++ + P RS GY G+++ISWR+ K +T++ SS+ A
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 64.7 bits (156), Expect = 1e-10
Identities = 42/107 (39%), Positives = 56/107 (52%)
Frame = -2
Query: 252 ARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAFLHGTLDEE 311
+R++AKG L F P+ K T+ LL++ A + +L LDV AFL G L E+
Sbjct: 593 SRILAKGRI------LL*NFVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVED 432
Query: 312 IYMRLPPGVSSPRPNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGY 358
IYM P G S V L+KS+YGLKQ RQ +L A G+
Sbjct: 431 IYMHQPEGFS*EVGKMVGKLKKSMYGLKQGPRQCI*SLKALCTRKGF 291
>BG648737 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberosum}, partial
(10%)
Length = 804
Score = 62.8 bits (151), Expect = 4e-10
Identities = 28/73 (38%), Positives = 41/73 (55%)
Frame = +3
Query: 162 HPLSSVLSYTNLSPKHYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLI 221
HP+ +SY LSP H AF +N+ P + ++ W+ A+ E+ AL +N TW +
Sbjct: 585 HPICKFISYDKLSPHHLAFVSNLDKIQIPNSVHESLTNP*WKKAILGEIHALEKNETWEL 764
Query: 222 VDLPPTKKPIGCK 234
DLP K P+GCK
Sbjct: 765 SDLPSGKHPMGCK 803
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 62.4 bits (150), Expect = 6e-10
Identities = 32/73 (43%), Positives = 41/73 (55%)
Frame = +2
Query: 620 LFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLP 679
L CD SA ++ NP +H R KHI +D H VR +Q G + + V T DQLAD TKPL
Sbjct: 23 LRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLADCLTKPLS 202
Query: 680 PSLFPTFVLKLGL 692
S K+G+
Sbjct: 203 KSRHQLLRNKIGV 241
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,518,147
Number of Sequences: 36976
Number of extensions: 371379
Number of successful extensions: 2190
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 2123
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2173
length of query: 694
length of database: 9,014,727
effective HSP length: 103
effective length of query: 591
effective length of database: 5,206,199
effective search space: 3076863609
effective search space used: 3076863609
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0260.2


