

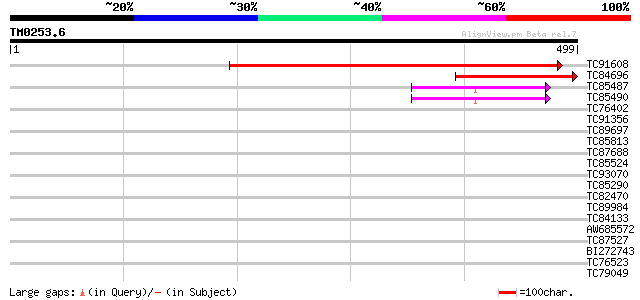
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.6
(499 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91608 similar to GP|14334894|gb|AAK59625.1 unknown protein {Ar... 510 e-145
TC84696 similar to GP|14334894|gb|AAK59625.1 unknown protein {Ar... 209 2e-54
TC85487 homologue to SP|P49688|RS2_ARATH 40S ribosomal protein S... 44 1e-04
TC85490 similar to GP|18146720|dbj|BAB82426. ribosomal protein S... 44 1e-04
TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F2... 34 0.11
TC91356 similar to GP|20151585|gb|AAM11152.1 LD24077p {Drosophil... 34 0.15
TC89697 similar to GP|4886756|gb|AAD32031.1| cellulose synthase ... 32 0.43
TC85813 homologue to GP|6503253|gb|AAC09468.2| putative NADPH-cy... 32 0.43
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 32 0.74
TC85524 similar to SP|P93508|CRTC_RICCO Calreticulin precursor. ... 31 1.3
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 30 1.6
TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsi... 30 1.6
TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical prot... 29 4.8
TC89984 similar to GP|16604601|gb|AAL24093.1 unknown protein {Ar... 29 4.8
TC84133 weakly similar to GP|22093574|dbj|BAC06871. contains EST... 29 4.8
AW685572 similar to GP|22830607|dbj PHD-finger family homeodomai... 29 4.8
TC87527 similar to GP|21689681|gb|AAM67462.1 unknown protein {Ar... 28 6.3
BI272743 homologue to GP|9279730|dbj formin-like protein {Arabid... 28 8.2
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 28 8.2
TC79049 weakly similar to PIR|T10798|T10798 pherophorin-S - Volv... 28 8.2
>TC91608 similar to GP|14334894|gb|AAK59625.1 unknown protein {Arabidopsis
thaliana}, partial (41%)
Length = 888
Score = 510 bits (1313), Expect = e-145
Identities = 249/293 (84%), Positives = 269/293 (90%)
Frame = +2
Query: 194 MNRLKVVTDQKDKFILQNKLNRALRLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQNAG 253
MNRLKVVT+QKD+FILQ+KLNRALRLVQWKEA+DPDNP NYG+IQ EQ G D L+ +G
Sbjct: 2 MNRLKVVTEQKDRFILQHKLNRALRLVQWKEAYDPDNPANYGLIQQEQGGANMDTLEQSG 181
Query: 254 FEKEKQTIQGQGDAADDDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAELEYTFGKKGK 313
FEKE + QG DAAD+DDE EFDDMKEKDNI+LAKLD IDK+LEEKLAELEYTFG+KGK
Sbjct: 182 FEKEDKPAQGDEDAADNDDEVEFDDMKEKDNILLAKLDVIDKRLEEKLAELEYTFGRKGK 361
Query: 314 VLEEEIRDLAEERNELTEKKRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNY 373
LEEEI+DLAEERNELTEKKRRPL+RKGFDVKLI+VNRTCKVTKGGQ+ KYTAMLACGNY
Sbjct: 362 ALEEEIKDLAEERNELTEKKRRPLFRKGFDVKLIDVNRTCKVTKGGQIFKYTAMLACGNY 541
Query: 374 NGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPT 433
NGVVGFAKAKGP P AL KAYEKCFQNLH++ERHEEHTIAHAIQTS+KKTKVYLWPA T
Sbjct: 542 NGVVGFAKAKGPKAPDALSKAYEKCFQNLHYIERHEEHTIAHAIQTSYKKTKVYLWPAST 721
Query: 434 ATGMKAGRIVQTILHLAGFKNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQ 486
ATGMKAGRIVQTILHLAGFKNVKSKVVG RNPHNTVKAVFKALNAIET +DVQ
Sbjct: 722 ATGMKAGRIVQTILHLAGFKNVKSKVVGQRNPHNTVKAVFKALNAIETQKDVQ 880
>TC84696 similar to GP|14334894|gb|AAK59625.1 unknown protein {Arabidopsis
thaliana}, partial (20%)
Length = 756
Score = 209 bits (532), Expect = 2e-54
Identities = 100/107 (93%), Positives = 105/107 (97%)
Frame = +1
Query: 393 KAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGF 452
+AYEKCFQNLH++ERHEEHTIAHAIQTS+KKTKVYLWPA TATGMKAGRIVQTILHLAGF
Sbjct: 64 QAYEKCFQNLHYIERHEEHTIAHAIQTSYKKTKVYLWPASTATGMKAGRIVQTILHLAGF 243
Query: 453 KNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKYLL 499
KNVKSKVVG RNPHNTVKAVFKALNAIETP+DVQEKFGRTVVEKYLL
Sbjct: 244 KNVKSKVVGQRNPHNTVKAVFKALNAIETPKDVQEKFGRTVVEKYLL 384
>TC85487 homologue to SP|P49688|RS2_ARATH 40S ribosomal protein S2.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (88%)
Length = 1114
Score = 43.9 bits (102), Expect = 1e-04
Identities = 36/131 (27%), Positives = 57/131 (43%), Gaps = 8/131 (6%)
Frame = +1
Query: 354 KVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERH----- 408
K T+ GQ ++ A + G+ NG VG V A++ A ++ V R
Sbjct: 379 KQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNK 558
Query: 409 --EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGS-RNP 465
+ HT+ + V + PAP +G+ A R+ + +L AG +V + GS +
Sbjct: 559 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 738
Query: 466 HNTVKAVFKAL 476
N VKA F L
Sbjct: 739 GNFVKATFDCL 771
>TC85490 similar to GP|18146720|dbj|BAB82426. ribosomal protein S2
{Arabidopsis thaliana}, partial (89%)
Length = 1252
Score = 43.9 bits (102), Expect = 1e-04
Identities = 36/131 (27%), Positives = 57/131 (43%), Gaps = 8/131 (6%)
Frame = +1
Query: 354 KVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERH----- 408
K T+ GQ ++ A + G+ NG VG V A++ A ++ V R
Sbjct: 358 KQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNK 537
Query: 409 --EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGS-RNP 465
+ HT+ + V + PAP +G+ A R+ + +L AG +V + GS +
Sbjct: 538 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 717
Query: 466 HNTVKAVFKAL 476
N VKA F L
Sbjct: 718 GNFVKATFDCL 750
>TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F23H14.7
{Arabidopsis thaliana}, partial (43%)
Length = 1449
Score = 34.3 bits (77), Expect = 0.11
Identities = 33/135 (24%), Positives = 59/135 (43%), Gaps = 14/135 (10%)
Frame = +3
Query: 65 AQVEKDRLREKNERMRAGLDT-ADIDAEGEED------------YMGVGPLIEKLEKEKL 111
A+ ++ +L+ + R+G DT +D D++ E + + + KEK
Sbjct: 783 AEAKEKKLKTAKHKKRSGSDTDSDRDSDDERKRASKRSHRKHRKHSHIDSGDHEKRKEKS 962
Query: 112 KECPELMRYEEPTDSDSDEDEYEA-SQKKFDDFERKFKRHQDLLKNFTDADTLDDAFKWM 170
+ R E +D SDE E + +K+ +RK R QD N + +D+ D
Sbjct: 963 SKWKTKKRSSESSDFSSDESESSSEEEKRRKKKQRKKIRDQDSRSNSSGSDSYAD----- 1127
Query: 171 NRIDKFEDKHFRLRP 185
++K + +H R RP
Sbjct: 1128-EVNKRKRRHRRRRP 1169
>TC91356 similar to GP|20151585|gb|AAM11152.1 LD24077p {Drosophila
melanogaster}, partial (71%)
Length = 698
Score = 33.9 bits (76), Expect = 0.15
Identities = 30/124 (24%), Positives = 53/124 (42%), Gaps = 7/124 (5%)
Frame = +2
Query: 345 KLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHF 404
+++ + K T+ GQ ++ A++ G+ G VG V A++ A ++
Sbjct: 326 EVMKIKPVQKQTRAGQRTRFKAIVLIGDSEGHVGLGIKTSKEVATAIRAAIIIAKLSVIP 505
Query: 405 VERHEEHT---IAHAIQTSFK----KTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKS 457
V T + H++ T V L PAP TG+ A V+ +L LAG ++ +
Sbjct: 506 VRLGYWGTNLGVPHSLPTKESGKCGSVTVRLIPAPRGTGLVASPAVKRLLQLAGVEDAYT 685
Query: 458 KVVG 461
G
Sbjct: 686 SSAG 697
>TC89697 similar to GP|4886756|gb|AAD32031.1| cellulose synthase catalytic
subunit {Arabidopsis thaliana}, partial (26%)
Length = 1011
Score = 32.3 bits (72), Expect = 0.43
Identities = 21/69 (30%), Positives = 34/69 (48%), Gaps = 7/69 (10%)
Frame = +2
Query: 101 PLIEKLEKEKLKECPEL-MRYEE-----PTDSDSDEDEYEASQKKFDDFERKFK-RHQDL 153
P E KE + CP+ RY+ + D DE++ + +++F E K+K HQD
Sbjct: 200 PCYEYERKEGSQNCPQCHTRYKRIKGSPRVEGDEDEEDVDDIEQEFKMEEEKYKLMHQDN 379
Query: 154 LKNFTDADT 162
+ + D DT
Sbjct: 380 MNSIDDDDT 406
>TC85813 homologue to GP|6503253|gb|AAC09468.2| putative NADPH-cytochrome
P450 reductase {Pisum sativum}, complete
Length = 2581
Score = 32.3 bits (72), Expect = 0.43
Identities = 45/193 (23%), Positives = 69/193 (35%), Gaps = 41/193 (21%)
Frame = +3
Query: 119 RYEEPTDSDSDEDEYEASQKKFDDFERKFKRHQDLLKNFTDADTLDDA---FKWMNRIDK 175
RYE+ D D+Y A ++++ +K L + D + D+A +KW +
Sbjct: 456 RYEKAKFKVVDMDDYAADDDEYEEKLKKETMALFFLATYGDGEPTDNAARFYKWFEEFEG 635
Query: 176 FEDKHFRLRPEYRVIG------ELMNRLKVVTDQKDKFILQNKLNRALRL---------- 219
ED L +Y V G E N++ + D K +L+ NR + +
Sbjct: 636 EEDSFKNL--QYGVFGLGNRQYEHFNKVAKIVDDK---LLEKGGNRLVPVGLGDDDQCIE 800
Query: 220 ---VQWKEAFDP-------DNPD------------NYGVIQHEQLGPTSDALQNAGFEKE 257
WKE P D D Y V+ +QL T D E
Sbjct: 801 DDFTAWKEELWPALDQLLRDEDDATVATPYTASVLEYRVVIRDQLDATVD---------E 953
Query: 258 KQTIQGQGDAADD 270
K+ + G G A D
Sbjct: 954 KKQLNGNGHAVVD 992
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 31.6 bits (70), Expect = 0.74
Identities = 43/191 (22%), Positives = 77/191 (39%), Gaps = 9/191 (4%)
Frame = +2
Query: 104 EKLEKEKLKECPELMRYEEPTDSDSDEDEYEASQKK--------FDDFERKFKRHQDLLK 155
+KLEK+K E E + + D D+D++E +KK DDFE + ++
Sbjct: 341 KKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDDEDEE--- 511
Query: 156 NFTDADTLDDAFKWMNRIDKFEDKHFRLRPEYRVIGELMNRLKVVTDQKDKFILQNKLNR 215
D D +D +K + K + ++ I EL L+ D +K
Sbjct: 512 GSEDEDDEED--------EKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEK--------- 640
Query: 216 ALRLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQNAG-FEKEKQTIQGQGDAADDDDEQ 274
+ ++ D D+ ++ D L+ AG FE + D DDDD++
Sbjct: 641 ----GEERDEADEDSEED-------------DELEKAGEFEMD--------DEDDDDDDE 745
Query: 275 EFDDMKEKDNI 285
E ++ ++ N+
Sbjct: 746 EDEEAEDMGNV 778
>TC85524 similar to SP|P93508|CRTC_RICCO Calreticulin precursor. [Castor bean]
{Ricinus communis}, partial (93%)
Length = 1699
Score = 30.8 bits (68), Expect = 1.3
Identities = 22/60 (36%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Frame = +1
Query: 107 EKEKLKECPELMRYEEPTDSDSDEDEYEASQKKFD-DFERKFKRHQDLLKNFTDADTLDD 165
E EK KE E ++P DSD++EDE +A++ D D E K + +D + T+ D + D
Sbjct: 1174 EAEKKKEEEETK--DDPVDSDAEEDEEDANEVSHDSDDESKAEAGED--SDETNKDDVHD 1341
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 30.4 bits (67), Expect = 1.6
Identities = 24/120 (20%), Positives = 51/120 (42%)
Frame = +1
Query: 245 TSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAEL 304
+ D ++ FE ++ + + + DDDD+ + +D + L+A DK+L+ K
Sbjct: 223 SDDEIEEMDFEDDEDEDEDEDEDDDDDDDDDDEDDDDDGFAEPTDLNAKDKRLKSKTVPD 402
Query: 305 EYTFGKKGKVLEEEIRDLAEERNELTEKKRRPLYRKGFDVKLINVNRTCKVTKGGQVVKY 364
++ K E+ +R L +++ K +R + + N + K + KY
Sbjct: 403 R----QQEKEKEKGVRSLNNGQSKRLPKSQR--------IASLQENNSAKFRRNSMEKKY 546
>TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsis
thaliana}, partial (32%)
Length = 1558
Score = 30.4 bits (67), Expect = 1.6
Identities = 26/100 (26%), Positives = 45/100 (45%), Gaps = 6/100 (6%)
Frame = +3
Query: 68 EKDRLREKNERMRAGLDTADIDAEGEEDYMGVGPLIEKLEKEKLKECPELMRYEEPTDSD 127
EK+R RE+ E+ + + + + E+ EK EKE+ +E + + +SD
Sbjct: 735 EKERKREE-EKAKKEKEREEKEKRKEK---------EKKEKEREREKEKSKERHKKDESD 884
Query: 128 SDEDE------YEASQKKFDDFERKFKRHQDLLKNFTDAD 161
SD + Y +KK D ERK +R + D++
Sbjct: 885 SDNQDMTDGHGYREEKKKEKDKERKHRRRHQSSMDDVDSE 1004
>TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (8%)
Length = 1064
Score = 28.9 bits (63), Expect = 4.8
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 7/73 (9%)
Frame = +2
Query: 269 DDDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAELEYTF---GKKGKVLEEEI----RD 321
DDDD+ + DD + D+ D L+EKL +E +F K +V E + RD
Sbjct: 35 DDDDDDDDDDDDDDDDDDDMSTDKSFSTLKEKLVLVEKSFEDIRSKTQVEERRLQSIKRD 214
Query: 322 LAEERNELTEKKR 334
+ E +L KK+
Sbjct: 215 IEECCEDLENKKK 253
>TC89984 similar to GP|16604601|gb|AAL24093.1 unknown protein {Arabidopsis
thaliana}, partial (32%)
Length = 1303
Score = 28.9 bits (63), Expect = 4.8
Identities = 25/104 (24%), Positives = 48/104 (46%), Gaps = 3/104 (2%)
Frame = +3
Query: 247 DALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKDNIILAKLDAIDKKLE---EKLAE 303
+ L+ E E + G D +E + ++++++L LDA ++LE ++ E
Sbjct: 528 EELRRKSLEMEMKLKSESGGNVGQDLTKE--SIVQQNDVLLQDLDAAREQLEILSKQYGE 701
Query: 304 LEYTFGKKGKVLEEEIRDLAEERNELTEKKRRPLYRKGFDVKLI 347
LE KVL +E++ L + EL ++ + K KL+
Sbjct: 702 LEAKSKADIKVLVKEVKSLRSSQTELKKELSESIKEKYEAEKLL 833
>TC84133 weakly similar to GP|22093574|dbj|BAC06871. contains ESTs
AU030377(E50973) AU030594(E51282)
AU030595(E51282)~similar to Arabidopsis thaliana,
partial (20%)
Length = 595
Score = 28.9 bits (63), Expect = 4.8
Identities = 11/23 (47%), Positives = 17/23 (73%)
Frame = +2
Query: 263 GQGDAADDDDEQEFDDMKEKDNI 285
G G DD+D +E D+++EKDN+
Sbjct: 98 GSGTPEDDNDIEERDNVEEKDNV 166
>AW685572 similar to GP|22830607|dbj PHD-finger family homeodomain protein
{Oryza sativa (japonica cultivar-group)}, partial (3%)
Length = 508
Score = 28.9 bits (63), Expect = 4.8
Identities = 15/42 (35%), Positives = 21/42 (49%)
Frame = +3
Query: 126 SDSDEDEYEASQKKFDDFERKFKRHQDLLKNFTDADTLDDAF 167
S SDE EY ++ +KF+D H+D D+ DD F
Sbjct: 36 SSSDESEYASASEKFED-----PGHEDQYMGLPSEDSEDDDF 146
>TC87527 similar to GP|21689681|gb|AAM67462.1 unknown protein {Arabidopsis
thaliana}, partial (96%)
Length = 1582
Score = 28.5 bits (62), Expect = 6.3
Identities = 14/42 (33%), Positives = 22/42 (52%)
Frame = +2
Query: 454 NVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
++ + + R+P +KA FKA E P+ QEK G T +
Sbjct: 1184 SIDNNLPPDRHPERRLKASFKAFEEAELPKLKQEKPGLTYTQ 1309
>BI272743 homologue to GP|9279730|dbj formin-like protein {Arabidopsis
thaliana}, partial (2%)
Length = 370
Score = 28.1 bits (61), Expect = 8.2
Identities = 25/92 (27%), Positives = 41/92 (44%), Gaps = 1/92 (1%)
Frame = +1
Query: 58 RIVQDLLAQVEKDRLREKNERMRA-GLDTADIDAEGEEDYMGVGPLIEKLEKEKLKECPE 116
++ + + + E++ +++ E + D D D E DY+ LIE+ E E
Sbjct: 106 KVEKIIFIEEEQEEMKDVEEEVELFDADDEDGDKGSEIDYI----LIEENNIE------E 255
Query: 117 LMRYEEPTDSDSDEDEYEASQKKFDDFERKFK 148
EE + +S E KKF+DF RK K
Sbjct: 256 EEHEEEEEEEESSVLSTEELNKKFEDFIRKMK 351
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 28.1 bits (61), Expect = 8.2
Identities = 23/78 (29%), Positives = 32/78 (40%), Gaps = 7/78 (8%)
Frame = +3
Query: 74 EKNERMRAGLDTADIDAEGEEDYMGVGPLIEKLEKEKLKECPELMRYEEPTDSDSDEDEY 133
+K+++ + D D D+ +ED V E E E E + SDSDE E
Sbjct: 243 KKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEVSESESDSSDDEDKMNVDKDGSDSDESEE 422
Query: 134 EAS-------QKKFDDFE 144
E+ QKK D E
Sbjct: 423 ESEDEPSKTPQKKIKDVE 476
>TC79049 weakly similar to PIR|T10798|T10798 pherophorin-S - Volvox carteri,
partial (6%)
Length = 957
Score = 28.1 bits (61), Expect = 8.2
Identities = 26/109 (23%), Positives = 46/109 (41%), Gaps = 5/109 (4%)
Frame = -3
Query: 41 LSSSYSGKSLYSTSSDARI--VQDLLAQVEKDRLREKNERMRAGLDTADIDAEGEEDYMG 98
L G S+AR+ ++ + +E + E+N+R+R + D E + + G
Sbjct: 862 LVKEVEGLKKVKAESEARVKDLEKRIGVLEMKEIEERNKRIRVEEELRDTIGEKDREIDG 683
Query: 99 VGPLIEKLEK---EKLKECPELMRYEEPTDSDSDEDEYEASQKKFDDFE 144
+E+LEK EK E + + + S E S++K FE
Sbjct: 682 FRNKVEELEKVGAEKKDEAGDWL-----NEKLSYEKALRESEEKAKGFE 551
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,049,802
Number of Sequences: 36976
Number of extensions: 139984
Number of successful extensions: 895
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 871
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 892
length of query: 499
length of database: 9,014,727
effective HSP length: 100
effective length of query: 399
effective length of database: 5,317,127
effective search space: 2121533673
effective search space used: 2121533673
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0253.6


