

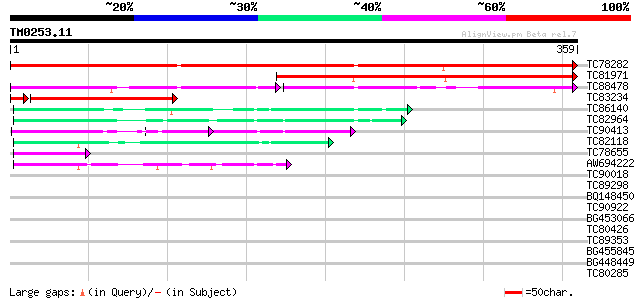
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.11
(359 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78282 weakly similar to PIR|B96836|B96836 unknown protein T21F... 478 e-135
TC81971 similar to PIR|B96836|B96836 unknown protein T21F11.23 [... 303 7e-83
TC88478 weakly similar to GP|20258927|gb|AAM14179.1 unknown prot... 122 3e-53
TC83234 122 6e-29
TC86140 similar to GP|9757759|dbj|BAB08240.1 emb|CAB55405.1~gene... 64 9e-11
TC82964 similar to GP|22655058|gb|AAM98120.1 predicted protein {... 58 6e-09
TC90413 similar to GP|21593314|gb|AAM65263.1 unknown {Arabidopsi... 46 3e-05
TC82118 similar to PIR|H86424|H86424 unknown protein [imported] ... 44 7e-05
TC78655 similar to PIR|B96698|B96698 unknown protein F12B7.3 [im... 42 4e-04
AW694222 weakly similar to PIR|T47927|T47 hypothetical protein T... 41 6e-04
TC90018 weakly similar to PIR|B84607|B84607 hypothetical protein... 40 0.001
TC89298 similar to PIR|B96698|B96698 unknown protein F12B7.3 [im... 38 0.005
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 36 0.026
TC90922 similar to PIR|F96594|F96594 unknown protein 58496-6030... 33 0.17
BG453066 32 0.29
TC80426 similar to GP|18377843|gb|AAL67108.1 AT5g42350/MDH9_4 {A... 32 0.38
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 32 0.38
BG455845 31 0.84
BG448449 weakly similar to GP|5903055|gb|A May be a pseudogene o... 30 1.4
TC80285 similar to GP|7300092|gb|AAF55261.1| srp gene product {D... 30 1.9
>TC78282 weakly similar to PIR|B96836|B96836 unknown protein T21F11.23
[imported] - Arabidopsis thaliana, partial (61%)
Length = 1657
Score = 478 bits (1230), Expect = e-135
Identities = 228/362 (62%), Positives = 280/362 (76%), Gaps = 3/362 (0%)
Frame = +2
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
MELISGLPEDVARDCLIRVSY+QFP V + CKGW TEI + E+ R+RR TGN+QK+LV
Sbjct: 212 MELISGLPEDVARDCLIRVSYQQFPTVASVCKGWKTEIHTVEYHRQRRRTGNSQKVLVMV 391
Query: 61 QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGY 120
QAR + + K NPVYRLSVFEPETG WSEL PP ++SG LP+ CQ+A VGY
Sbjct: 392 QARIEPNKTETGSTKRLVNPVYRLSVFEPETGFWSELPAPPGYNSG--LPVMCQVACVGY 565
Query: 121 ELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHD 180
+LVV+GG DPE+WKASNSVF+YNFLSA WR G MPGGPRTFF C+SD +T++VAGGHD
Sbjct: 566 DLVVLGGLDPETWKASNSVFVYNFLSAKWRCGTQMPGGPRTFFGCSSDDRQTIFVAGGHD 745
Query: 181 EEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERS 240
+EKNAL+SALAYDV D WV LP+M+ ERDECKAVFRG G+ VVGGY TE QGRFERS
Sbjct: 746 DEKNALRSALAYDVVADVWVQLPEMSSERDECKAVFRG--GRFIVVGGYTTENQGRFERS 919
Query: 241 AEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGD---AMYMCRGGDVVALQGNTWQTVAKV 297
AE FD TW+WG VEEE+LDC TCP T +DGGD ++YMC G++V ++ +TWQ + +V
Sbjct: 920 AEAFDFVTWKWGQVEEEYLDCATCPMTLVDGGDEEESVYMCCNGELVMMRAHTWQKMGRV 1099
Query: 298 PSEIRNVACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYCLL 357
P EI NVA +GA++ +++IGSSG+GE HMG++ D+ + W KL P + GHVQ+ C+L
Sbjct: 1100PDEICNVAYVGAFDGFVVVIGSSGYGEVHMGYVFDVNNNNWRKLDCPDGFKGHVQTGCVL 1279
Query: 358 EI 359
EI
Sbjct: 1280EI 1285
>TC81971 similar to PIR|B96836|B96836 unknown protein T21F11.23 [imported] -
Arabidopsis thaliana, partial (13%)
Length = 721
Score = 303 bits (776), Expect = 7e-83
Identities = 146/196 (74%), Positives = 164/196 (83%), Gaps = 6/196 (3%)
Frame = +1
Query: 170 DRTVYVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVF---RGGAGKLRVV 226
+R VYVAGGHDEEKNALKSA AYDV +D W+PLPDMARERDECK VF G+G ++VV
Sbjct: 1 ERMVYVAGGHDEEKNALKSAFAYDVVDDMWIPLPDMARERDECKVVFCAKDNGSGTIKVV 180
Query: 227 GGYCTEMQGRFERSAEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGDA---MYMCRGGDV 283
GGY TEMQGRFERSAEEF V TW+WGPVEEEFLD TCPRTC+DG D MYMC+G DV
Sbjct: 181 GGYRTEMQGRFERSAEEFGVATWRWGPVEEEFLDDATCPRTCVDGCDLERKMYMCKGDDV 360
Query: 284 VALQGNTWQTVAKVPSEIRNVACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVS 343
VAL G TWQ VAKVP EIRNVAC+GAW +LLLIGSSGFGEP+M F+LDLKSG W+KL +
Sbjct: 361 VALDGETWQVVAKVPREIRNVACVGAWVDALLLIGSSGFGEPYMSFVLDLKSGVWSKLEN 540
Query: 344 PQDYTGHVQSYCLLEI 359
P+++TGHVQS CLLEI
Sbjct: 541 PENFTGHVQSGCLLEI 588
>TC88478 weakly similar to GP|20258927|gb|AAM14179.1 unknown protein
{Arabidopsis thaliana}, partial (8%)
Length = 1147
Score = 122 bits (307), Expect(2) = 3e-53
Identities = 69/178 (38%), Positives = 95/178 (52%), Gaps = 7/178 (3%)
Frame = +2
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
+ELI GLP ++ +CL R+S C W S EF R+ TG+T+K+
Sbjct: 113 IELIPGLPSELGLECLTRLSNSSHRVALRVCNQWRRLFLSDEFYSHRKNTGHTRKVACLV 292
Query: 61 QAR-------FDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFC 113
QA FD T +T P Y ++VF+PE+ +W + PE+ SG LP+FC
Sbjct: 293 QAHEQQPHSEFDKQT-------GSTPPSYDITVFDPESMSWDRVDPVPEYPSG--LPLFC 445
Query: 114 QIAGVGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDR 171
Q+ +LVVMGGWDP S++ +VF+Y+F W RG DMP R+FFA S DR
Sbjct: 446 QLTSCEGKLVVMGGWDPASYEPLTAVFVYDFRMNIWWRGKDMP-EKRSFFATGSGYDR 616
Score = 103 bits (258), Expect(2) = 3e-53
Identities = 71/190 (37%), Positives = 95/190 (49%), Gaps = 4/190 (2%)
Frame = +3
Query: 174 YVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEM 233
+VAGGHDE KNALK+A AYD DEW L M+++RDEC+ G G+ VV GY TE
Sbjct: 621 FVAGGHDENKNALKTAWAYDPKIDEWTMLAPMSQDRDECEGTVVG--GEFWVVSGYATES 794
Query: 234 QGRFERSAEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNTWQT 293
QG F+ SAE D+ + QW VE + + G CPR+C+D + G V
Sbjct: 795 QGMFDDSAEVLDIGSGQWRRVEGVW-EAGRCPRSCVD------IRENGKV---------- 923
Query: 294 VAKVPSEIRNVACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVS----PQDYTG 349
V +R C + GS G P+ +L++ G+ KL P + G
Sbjct: 924 ---VDPGLRIGVCSVRVGSRKWVTGSEYEGAPYGFYLVENDEGQNRKLNKISSVPDGFAG 1094
Query: 350 HVQSYCLLEI 359
VQS C +EI
Sbjct: 1095FVQSGCCVEI 1124
>TC83234
Length = 516
Score = 122 bits (305), Expect(2) = 6e-29
Identities = 60/94 (63%), Positives = 71/94 (74%), Gaps = 1/94 (1%)
Frame = +1
Query: 14 DCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQARFDSD-TCGGL 72
+CL+RVSY++FPAV KGW TEIQ+PEFRR RR TG+ QKILVT Q++FDS+ + GL
Sbjct: 232 ECLVRVSYQEFPAVATVSKGWQTEIQTPEFRRVRRSTGHAQKILVTVQSKFDSEKSKTGL 411
Query: 73 LVKATTNPVYRLSVFEPETGNWSELTMPPEFDSG 106
L KAT NPVY L+V E ETG WSEL M PE G
Sbjct: 412 LAKATANPVYNLNVLETETGIWSELPMGPELCEG 513
Score = 23.1 bits (48), Expect(2) = 6e-29
Identities = 10/12 (83%), Positives = 11/12 (91%)
Frame = +3
Query: 1 MELISGLPEDVA 12
MELIS LPED+A
Sbjct: 192 MELISCLPEDIA 227
>TC86140 similar to GP|9757759|dbj|BAB08240.1
emb|CAB55405.1~gene_id:MUF9.20~similar to unknown
protein {Arabidopsis thaliana}, partial (88%)
Length = 1825
Score = 63.9 bits (154), Expect = 9e-11
Identities = 66/258 (25%), Positives = 98/258 (37%), Gaps = 5/258 (1%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
L+ GL +DVA +CL V +++ K ++ I S R+ G + ++
Sbjct: 333 LLPGLIDDVALNCLAWVGRSNHASLSCINKRYSRLISSGYLYGLRKELGVVEHLVYLV-- 506
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPP-----EFDSGSGLPMFCQIAG 117
D G + F+P G W L P L + C++
Sbjct: 507 ---CDPRGWV-------------AFDPTIGRWMALPKIPCDECFNHADKESLAVGCELLV 638
Query: 118 VGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAG 177
G EL+ W Y+ + W + +M PR F +S VAG
Sbjct: 639 FGRELMEFAIWK------------YSLICRRWLKCPEM-NQPRCLFG-SSSLGSIAIVAG 776
Query: 178 GHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRF 237
G D+ N LKSA YD A+ W LP+M R C + F GK V+GG +
Sbjct: 777 GSDKYGNVLKSAELYDSASCTWELLPNMHTPRRLCSSFFMD--GKFYVIGGMSSTTVSL- 947
Query: 238 ERSAEEFDVDTWQWGPVE 255
EE+D+ T W +E
Sbjct: 948 -TCGEEYDLSTRSWRKIE 998
>TC82964 similar to GP|22655058|gb|AAM98120.1 predicted protein {Arabidopsis
thaliana}, partial (73%)
Length = 1244
Score = 57.8 bits (138), Expect = 6e-09
Identities = 59/249 (23%), Positives = 101/249 (39%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
L+ + D + CL R S + ++ + + + I+S E R RR+ G + + + A
Sbjct: 153 LLPRMNRDSSITCLSRCSRSDYGSLASLNQSFRNIIRSGELYRWRRLNGIIEHWIYFSCA 332
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYEL 122
+ + ++P W L P + + + VG EL
Sbjct: 333 LLEWEA------------------YDPIRERWMHL--PRMASNECFMCSDKESLAVGTEL 452
Query: 123 VVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHDEE 182
+V G S+ ++ Y+ L+ +W G M PR F S ++ + +AGG D +
Sbjct: 453 LVFG-----RELQSHVIYRYSLLTNSWESGMRM-NFPRCLFGSASLREIAI-LAGGCDSD 611
Query: 183 KNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERSAE 242
+ L SA Y+ N W LP+M + R C VF G V+GG + E
Sbjct: 612 GHILDSAELYNSENQTWETLPNMIKPRKMCSGVFMD--GNFYVIGG-IGGRDSKLLTCGE 782
Query: 243 EFDVDTWQW 251
E+++ T W
Sbjct: 783 EYNLQTRTW 809
>TC90413 similar to GP|21593314|gb|AAM65263.1 unknown {Arabidopsis
thaliana}, partial (64%)
Length = 1104
Score = 45.8 bits (107), Expect = 3e-05
Identities = 36/134 (26%), Positives = 57/134 (41%), Gaps = 1/134 (0%)
Frame = +1
Query: 87 FEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYELVVMGGWDPESWKASNSVFIYNFLS 146
F +G L + P+ +S C + V L+++ A++ V+ Y+ +
Sbjct: 532 FPQRSGTLLTLVLYPQLES-------CLSSAVEVMLLILRPVITMGCFATDEVWSYDPII 690
Query: 147 ATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHDEEKNALKSALAYDVANDEWVPLPDMA 206
W A M PR+ FAC + V VAGG + + A YD D W P+PD+
Sbjct: 691 QQWAPRASMLV-PRSMFACCVLNGKIV-VAGGFTSCRKTISQAEMYDPEKDVWTPMPDLH 864
Query: 207 R-ERDECKAVFRGG 219
R + C + GG
Sbjct: 865 RTQNSACSGIVIGG 906
Score = 43.5 bits (101), Expect = 1e-04
Identities = 35/128 (27%), Positives = 55/128 (42%)
Frame = +3
Query: 2 ELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQ 61
ELI GLP+ VA CL RV + P + + W I SPE + R+ +++ +L
Sbjct: 285 ELIEGLPDAVAIRCLARVPFYLHPKLEVVSRSWQAAIHSPELSKIRQEVASSEDLLCV-- 458
Query: 62 ARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYE 121
FD + V++L ++P W +T+P L F ++ G
Sbjct: 459 CAFDPEN------------VWQL--YDPLRDLW--ITLPVLPSKIRHLAHFGAVSAAGKL 590
Query: 122 LVVMGGWD 129
V+ GG D
Sbjct: 591 FVIGGGSD 614
>TC82118 similar to PIR|H86424|H86424 unknown protein [imported] -
Arabidopsis thaliana, partial (61%)
Length = 840
Score = 44.3 bits (103), Expect = 7e-05
Identities = 51/205 (24%), Positives = 79/205 (37%), Gaps = 2/205 (0%)
Frame = +2
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPE--FRRRRRVTGNTQKILVTA 60
LI GLP+DVA +CL+R+ + + A CK W+ + + E F R+++ + V A
Sbjct: 227 LIPGLPDDVALNCLLRLPVQSHSSCRAVCKRWHMLLGNKERFFTNRKQMGFKDPWLFVFA 406
Query: 61 QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGY 120
+ C G + + V + +W + P D C
Sbjct: 407 YHK-----CTGKI---------QWQVLDLTHFSWHTIPAMPCKDKVCPHGFRCVSMPHDG 544
Query: 121 ELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHD 180
L V GG + + V Y W M R+FFA + D VY AGG+
Sbjct: 545 TLYVCGGMVSDVDCPLDLVLKYEITKNRWTVMNRMISA-RSFFA-SGVIDGMVYAAGGNS 718
Query: 181 EEKNALKSALAYDVANDEWVPLPDM 205
+ L SA D + W + +M
Sbjct: 719 TDLYELDSAEVLDPISGNWRAIANM 793
>TC78655 similar to PIR|B96698|B96698 unknown protein F12B7.3 [imported] -
Arabidopsis thaliana, partial (16%)
Length = 802
Score = 42.0 bits (97), Expect = 4e-04
Identities = 20/49 (40%), Positives = 28/49 (56%)
Frame = +1
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTG 51
++ GLP+DVA+ CL V FPA+ K W I+S EF R++ G
Sbjct: 625 ILPGLPDDVAKYCLALVPRSNFPAMGGVSKKWRLFIRSKEFVMVRKLAG 771
>AW694222 weakly similar to PIR|T47927|T47 hypothetical protein T20K12.250 -
Arabidopsis thaliana, partial (27%)
Length = 651
Score = 41.2 bits (95), Expect = 6e-04
Identities = 45/184 (24%), Positives = 76/184 (40%), Gaps = 8/184 (4%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPE---FRRRRRVTGNTQKILVT 59
LI GLP+D++ CL RV + + A K W + S E +RR+ ++ + I
Sbjct: 39 LICGLPDDISLLCLARVPRKYHSVLKAVSKRWKDLVCSEEWLYYRRKHKL--DETWIYAL 212
Query: 60 AQARFDSDTCGGLLVKATTNPVYRLSVFEPETG--NWSEL-TMPPEFDSGSGLPMFCQIA 116
+ + D C V +P + +W + +PP G+
Sbjct: 213 CRDKLDHVYC---------------YVLDPTSSRKSWKLIHGVPPHVMKRKGM----GFE 335
Query: 117 GVGYELVVMG--GWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVY 174
+G +L ++G GW + A++ V+ YN W A + R +FAC + D +Y
Sbjct: 336 ALGNKLFLLGGCGWSED---ATDEVYSYNASLNLWVEAASL-STARCYFACEA-MDEKLY 500
Query: 175 VAGG 178
GG
Sbjct: 501 AIGG 512
>TC90018 weakly similar to PIR|B84607|B84607 hypothetical protein At2g21950
[imported] - Arabidopsis thaliana, partial (55%)
Length = 965
Score = 40.4 bits (93), Expect = 0.001
Identities = 62/250 (24%), Positives = 95/250 (37%), Gaps = 6/250 (2%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LI LP+DVA +CL RV +T K ++ + S F R + +TQ IL +
Sbjct: 42 LIPSLPDDVAINCLARVPRSHHTTLTLVSKPIHSLLSSSLFFTARSLIPSTQHILYLS-- 215
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYEL 122
L ++T+ + L N L +PP G A + +++
Sbjct: 216 ---------LRTRSTSLQFFTLH------NNHRLLPLPPLPSPTIG----SAYAVIHHKI 338
Query: 123 VVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGG--HD 180
++GG + S V+I + W G M FA D +YV GG D
Sbjct: 339 YLIGG--SVNDVPSRHVWILDCRFHRWLPGPSMRVARE--FAAAGVIDGKIYVIGGCVPD 506
Query: 181 EEKNALKSALAYDVANDEW--VPLPDMARER-DECKAVFRGGAGKLRVVGGYCTE-MQGR 236
+ + +D N+ W VP P RE+ AV G + GG + G
Sbjct: 507 NFSRSANWSEVFDPVNNRWESVPSPPEIREKWMHASAVVDGKVYAMADRGGVSFDPYNGA 686
Query: 237 FERSAEEFDV 246
+E E D+
Sbjct: 687 WESVGRELDI 716
Score = 28.1 bits (61), Expect = 5.5
Identities = 29/126 (23%), Positives = 47/126 (37%), Gaps = 2/126 (1%)
Frame = +3
Query: 173 VYVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTE 232
+Y+ GG + + + D W+P P M R+ A G K+ V+GG +
Sbjct: 336 IYLIGGSVNDVPS-RHVWILDCRFHRWLPGPSMRVAREFAAAGVIDG--KIYVIGGCVPD 506
Query: 233 MQGRFERSAEEFDV--DTWQWGPVEEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNT 290
R +E FD + W+ P E + +DG RGG
Sbjct: 507 NFSRSANWSEVFDPVNNRWESVPSPPEIREKWMHASAVVDGKVYAMADRGGVSFDPYNGA 686
Query: 291 WQTVAK 296
W++V +
Sbjct: 687 WESVGR 704
>TC89298 similar to PIR|B96698|B96698 unknown protein F12B7.3 [imported] -
Arabidopsis thaliana, partial (31%)
Length = 898
Score = 38.1 bits (87), Expect = 0.005
Identities = 24/66 (36%), Positives = 33/66 (49%)
Frame = +1
Query: 163 FACTSDQDRTVYVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGK 222
FAC ++ D VY+ GG+ + L S YD D+W + + R R C A G K
Sbjct: 34 FAC-AEVDGLVYIVGGYGVNGDNLSSVEMYDPDTDKWTLIESLRRPRWGCFAC--GFEDK 204
Query: 223 LRVVGG 228
L V+GG
Sbjct: 205 LYVMGG 222
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 35.8 bits (81), Expect = 0.026
Identities = 44/181 (24%), Positives = 78/181 (42%), Gaps = 13/181 (7%)
Frame = +3
Query: 7 LPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQARFDS 66
LP ++ + L R+ + CK W ++I P+F ++ NT+ + + ++
Sbjct: 3 LPFEIQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKHLRVSNTRHLFLLTFSKLSP 182
Query: 67 DTCGGLLVKATTNPVYRL-SVFEPETGNWSELTMP----PEFDS--GSGLPMFCQIAGVG 119
+ L++K+ Y L SVF T +++L P E DS GS + C +
Sbjct: 183 E----LVIKS-----YPLSSVFTEMTPTFTQLEYPLNNRDESDSMVGSCHGILCIQCNLS 335
Query: 120 YELVVMGGWDP---ESWKASNSVFIYN-FLSATWRRGADMPGGPRTFFA--CTSDQDRTV 173
+ ++ W+P + K + F N F++ T+ G D A CTS+ D V
Sbjct: 336 FPVL----WNPSIRKFTKLPSFEFPQNKFINPTYAFGYDHSSDTYKVVAVFCTSNIDNGV 503
Query: 174 Y 174
Y
Sbjct: 504 Y 506
>TC90922 similar to PIR|F96594|F96594 unknown protein 58496-60308
[imported] - Arabidopsis thaliana, partial (31%)
Length = 696
Score = 33.1 bits (74), Expect = 0.17
Identities = 17/53 (32%), Positives = 26/53 (48%)
Frame = +1
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQK 55
L+ GLP+D+A CLIRV + + CK W + F R+ G ++
Sbjct: 445 LLPGLPDDLAIACLIRVPRVEHRKLRLVCKRWYRLLSGNFFYSLRKSLGMAEE 603
>BG453066
Length = 655
Score = 32.3 bits (72), Expect = 0.29
Identities = 24/81 (29%), Positives = 39/81 (47%), Gaps = 7/81 (8%)
Frame = +1
Query: 135 ASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGG-----HDEEKNALKSA 189
+ N VF+Y + W+R D+P + CT + +YV+G HDE ++L
Sbjct: 427 SKNEVFLYTLGTDYWKRIDDIP------YYCTICRS-GLYVSGAVNWYVHDESXSSLYFI 585
Query: 190 LAYDVANDEWVPL--PDMARE 208
L+ D+ N+ + L PD E
Sbjct: 586 LSLDLXNESYQELYPPDFXDE 648
>TC80426 similar to GP|18377843|gb|AAL67108.1 AT5g42350/MDH9_4 {Arabidopsis
thaliana}, partial (14%)
Length = 844
Score = 32.0 bits (71), Expect = 0.38
Identities = 13/42 (30%), Positives = 20/42 (46%)
Frame = +1
Query: 7 LPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRR 48
LP+D+ CL+R+ CK W + +P F + RR
Sbjct: 637 LPDDILEMCLVRLPLTSLMNARLVCKKWRSLTTTPRFLQLRR 762
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 32.0 bits (71), Expect = 0.38
Identities = 12/45 (26%), Positives = 23/45 (50%)
Frame = +3
Query: 2 ELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRR 46
E+ + +PE++ + L+R+ CK W T I P+F ++
Sbjct: 90 EITADMPEEIIVEILLRLPVRSLLQFRCVCKLWKTLISDPQFAKK 224
>BG455845
Length = 671
Score = 30.8 bits (68), Expect = 0.84
Identities = 26/104 (25%), Positives = 45/104 (43%), Gaps = 7/104 (6%)
Frame = +3
Query: 127 GWDPESWKA-------SNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGH 179
GWD +W SV + + + + + GGP F C T V GH
Sbjct: 141 GWDKVAWPTYAPDCDQGGSVELDKTTAHSGKNSIKVTGGPNGF--CGHKFFGTDAVPEGH 314
Query: 180 DEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKL 223
+ +K+A A+D ++ ++ +PD A+ + K + GG K+
Sbjct: 315 VYVRTWMKTAKAFDDSHVTFITMPDAAQGKG--KHLRIGGQSKI 440
>BG448449 weakly similar to GP|5903055|gb|A May be a pseudogene of F6D8.29.
{Arabidopsis thaliana}, partial (12%)
Length = 679
Score = 30.0 bits (66), Expect = 1.4
Identities = 25/108 (23%), Positives = 46/108 (42%), Gaps = 7/108 (6%)
Frame = +3
Query: 7 LPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQARFDS 66
+P D++ D + ++ + T CK W ++P+F R +++R+D
Sbjct: 69 IPHDLSFDIVSKLPLKSLKRFTCVCKFWANLFENPQFMSVYR------NNFFLSRSRYDD 230
Query: 67 DTCGGLLVKATTNPVY------RLSVFEPET-GNWSELTMPPEFDSGS 107
LL+K T P Y ++ + +T N +L PP F+ S
Sbjct: 231 HQNSRLLLKVT--PAYGYGRVDKMFLLSGDTFENSVKLDWPPLFEEDS 368
>TC80285 similar to GP|7300092|gb|AAF55261.1| srp gene product {Drosophila
melanogaster}, partial (2%)
Length = 1055
Score = 29.6 bits (65), Expect = 1.9
Identities = 13/40 (32%), Positives = 21/40 (52%)
Frame = -2
Query: 7 LPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRR 46
LPE++ + + RV + CK WN+ I PEF ++
Sbjct: 997 LPEELIIEIISRVDSRNTLQLRPVCKRWNSLITDPEFVKK 878
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,441,745
Number of Sequences: 36976
Number of extensions: 185405
Number of successful extensions: 718
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 697
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 706
length of query: 359
length of database: 9,014,727
effective HSP length: 97
effective length of query: 262
effective length of database: 5,428,055
effective search space: 1422150410
effective search space used: 1422150410
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0253.11


