

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.10
(674 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
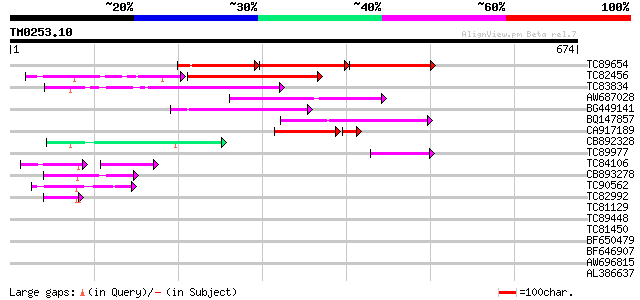
Score E
Sequences producing significant alignments: (bits) Value
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 105 1e-68
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 172 1e-51
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 154 1e-37
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 143 2e-34
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 126 3e-29
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 101 1e-21
CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.3... 79 4e-17
CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sin... 70 3e-12
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 58 1e-08
TC84106 similar to GP|9502168|gb|AAF88018.1| contains simlarity ... 40 1e-07
CB893278 weakly similar to PIR|T05645|T056 hypothetical protein ... 54 2e-07
TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 49 5e-06
TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D... 43 3e-04
TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 40 0.003
TC89448 similar to PIR|C84864|C84864 hypothetical protein At2g43... 40 0.004
TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 32 0.005
BF650479 homologue to PIR|T06643|T066 hypothetical protein T20K1... 38 0.011
BF646907 similar to GP|15983442|gb| At1g10240/F14N23_12 {Arabido... 35 0.12
AW696815 homologue to GP|11994736|dbj far-red impaired response ... 33 0.36
AL386637 weakly similar to GP|13325257|gb|A Similar to CG15432 g... 32 0.61
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 105 bits (263), Expect(3) = 1e-68
Identities = 48/98 (48%), Positives = 64/98 (64%)
Frame = +2
Query: 200 YIRNVRRKNLDVGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQL 259
++ + R++ L G Q V DY KR + EN FFYA+Q D D+ N+FW D R +Y
Sbjct: 83 HMSSTRQRTLGGGGHQ-VLDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSY 259
Query: 260 FGDVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCA 297
FGD + FDTTYKTN+Y +PFA F G N+H Q +L+GCA
Sbjct: 260 FGDTVIFDTTYKTNQYRVPFASFTGFNHHGQPVLFGCA 373
Score = 104 bits (260), Expect(3) = 1e-68
Identities = 44/107 (41%), Positives = 68/107 (63%)
Frame = +3
Query: 298 LLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMKAALAKVFPESRHRLCLWHIIKK 357
L+ +ESE S++WLFKTWL+A+ G+ P+SI TD D ++ A+A+V P +RHR C W I ++
Sbjct: 375 LILNESEPSYIWLFKTWLRAVSGRPPVSITTDLDPVIQVAVAQVLPPTRHRFCKWSIFRE 554
Query: 358 FPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFEEEWMRIMVEYNLKE 404
KLAH+Y F + K+C+ S +I FE W ++ Y + +
Sbjct: 555 NRSKLAHLYQSNPTFDNEFKKCVHESVTIDEFESCWRSLLERYYIMD 695
Score = 89.7 bits (221), Expect(3) = 1e-68
Identities = 42/102 (41%), Positives = 65/102 (63%)
Frame = +1
Query: 405 NEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDASTTLQEFVLKFEKA 464
NEW Q +Y R+ W+P+Y R +FF + +E +N FFD V+ASTTLQ V ++EKA
Sbjct: 697 NEWPQSMYNARQHWVPVYLRGSFFGEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEKA 876
Query: 465 VESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIFL 506
V + E E K ++E+ + + +L T S +E+ A LYT ++F+
Sbjct: 877 VSTWHERELKADFETSNSNPVLRTPSPMEKQAASLYTRKMFM 1002
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 172 bits (437), Expect(2) = 1e-51
Identities = 78/160 (48%), Positives = 104/160 (64%)
Frame = +2
Query: 212 GDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYK 271
GD A+ + R Q +NS F+ A+ D+ + NLFW DAR R +Y+ FG+VIT DTTY
Sbjct: 713 GDVDAIHSFFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDTTYL 892
Query: 272 TNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQD 331
T+KY +P PFVG+N+H Q+IL GCA+L + + WLF WL+ M G P IIT++D
Sbjct: 893 TSKYDLPLVPFVGVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECMHGHAPNGIITEED 1072
Query: 332 LAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSI 371
AMK A+ FP++RHR CLWHI+KK PE L +SI
Sbjct: 1073KAMKNAIEVAFPKARHRWCLWHIMKKVPEMLGKYSRNESI 1192
Score = 49.7 bits (117), Expect(2) = 1e-51
Identities = 50/199 (25%), Positives = 87/199 (43%), Gaps = 9/199 (4%)
Frame = +1
Query: 20 SSKDFTTSILDTSDIGGELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSK---- 75
S F+ +I+ + D +P GM F S E+V +Y ++ + GFGV SSK
Sbjct: 160 SDDKFSAAIVVSED------EEPRTGMTFRSEEEVIRYYMNYANRVGFGVTKISSKNADD 321
Query: 76 PKRAVLVCCNEGQHKVKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIK 135
K+ + CN + V S+ + T++T+ R+ A + + +T S+ ++
Sbjct: 322 GKKYFTLACNCARRYVSTSKNPSKQYLTSKTQ-----CRARLNACVALDGSSTISRIVL- 483
Query: 136 SFNNDHNHVMVSPKSVSYMRCHKKMSVAAKSLVEKFEEEGIPTGK----VAAIFND-GDS 190
+HNH + K Y R + + +E ++ GI + V N G+
Sbjct: 484 ----EHNHELGQTKG-RYFRLDESSGTHVERNLELNDQGGINVDRNLQSVGLEMNGCGNL 648
Query: 191 TFTNRDCWNYIRNVRRKNL 209
T DC N+++ VRR L
Sbjct: 649 TSGENDCRNFVQKVRRLRL 705
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 154 bits (389), Expect = 1e-37
Identities = 96/295 (32%), Positives = 150/295 (50%), Gaps = 10/295 (3%)
Frame = +3
Query: 42 PEIGMEFESIEKVREFYNSFDKKNGFGVRV------RSSKPKRAVLVCCNEGQHKVKISR 95
P + MEFES + +Y + K+ GF VRV R+SK K ++CC+ K R
Sbjct: 213 PALAMEFESYDDAYSYYICYAKEVGFCVRVKNSWFKRNSKEKYGAVLCCSSQGFK----R 380
Query: 96 TEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMR 155
T+++ + +T R+GC A +I + +W I +HNHV+ + +
Sbjct: 381 TKDVNNLRKET-------RTGCPA-MIRMKLVESQRWRICEVTLEHNHVLGA-------K 515
Query: 156 CHKKMSVAAKSLVEKFEEEG--IPTGKVAAIFNDGDSTFTN--RDCWNYIRNVRRKNLDV 211
HK + K+ + + EG I I +G+ + RD + + + NL
Sbjct: 516 IHKSIK---KNSLPSSDAEGKTIKVYHALVIDTEGNDNLNSNARDDRAFSKYSNKLNLRK 686
Query: 212 GDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYK 271
GD QA++++ R Q+ N NFFY + +++ + N WVDA+SR + F DVI FD TY
Sbjct: 687 GDTQAIYNFLCRMQLTNPNFFYLMDFNDEGHLRNALWVDAKSRAACGYFSDVIYFDNTYL 866
Query: 272 TNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISI 326
NKY +P VG+N+H QS+L GC LL E S+ WLF+TW++ + P +I
Sbjct: 867 VNKYEIPLVALVGINHHGQSVLLGCGLLAGEIIESYKWLFRTWIKCIPRCSPQTI 1031
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 143 bits (361), Expect = 2e-34
Identities = 67/186 (36%), Positives = 110/186 (59%)
Frame = +1
Query: 262 DVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGK 321
DV+ FDTTYK NKY+ P F G N+HSQ+I++G ALL+DE+ S+ W+ +L+ M K
Sbjct: 1 DVLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENK 180
Query: 322 KPISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIR 381
P +++TD D +M+ A+ +VFP++ HRLC WH+ K E + K++ F ++ +
Sbjct: 181 FPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENI-----KKTPFLEGFRKAMY 345
Query: 382 GSHSIQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESIN 441
+ + + FE+ W ++ + L+ N W+ Y + W Y R FF + TT + E+IN
Sbjct: 346 SNFTPEQFEDFWSELIQKNELEGNAWVIKTYANKSLWATAYLRDKFFGRIRTTSQCEAIN 525
Query: 442 AFFDSF 447
A ++
Sbjct: 526 AIVKTY 543
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 126 bits (316), Expect = 3e-29
Identities = 58/168 (34%), Positives = 98/168 (57%)
Frame = +3
Query: 192 FTNRDCWNYIRNVRRKNLDVGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDA 251
FT +D N +++ R+ + + + + C+ + ++ NF + D ++R+ N+ W A
Sbjct: 171 FTEKDVRNLLQSFRKLDPEE-ETLDLLRMCRNIKDKDPNFKFEYTLDANNRLENIAWSYA 347
Query: 252 RSRLSYQLFGDVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLF 311
S Y +FGD + FDTT++ + MP +VG+NN+ +GC LL+DE+ SF W
Sbjct: 348 SSIQLYDIFGDAVVFDTTHRLTAFDMPLGLWVGINNYGMPCFFGCVLLRDETVRSFSWAI 527
Query: 312 KTWLQAMGGKKPISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFP 359
K +L M GK P +I+TDQ++ +K AL+ P ++H C+W I+ KFP
Sbjct: 528 KAFLGFMNGKAPQTILTDQNICLKEALSAEMPMTKHAFCIWMIVAKFP 671
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 101 bits (251), Expect = 1e-21
Identities = 55/181 (30%), Positives = 93/181 (50%), Gaps = 1/181 (0%)
Frame = +1
Query: 323 PISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQ-SIFKRDMKRCIR 381
P +++TD + +K A+A PES+H C+WHI+ KF + + Q +K D R +
Sbjct: 4 PQTLLTDHNTWLKEAIAVEMPESKHAFCIWHILSKFSDWXYLLLGSQYDEWKADFHR-LY 180
Query: 382 GSHSIQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESIN 441
+ FE+ W +++ +Y L N+ + LY +R W + R FFAG+ + ++ESIN
Sbjct: 181 NLEMXEDFEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAGLTSXCQTESIN 360
Query: 442 AFFDSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYT 501
F F+ A + F+ + V+ A K+ + + + L TGS +E HA + T
Sbjct: 361 VFIQRFLSAQSQPXRFLQQVADIVDFNDRAGAKQXMQRKMQKXCLKTGSPIESHAATILT 540
Query: 502 P 502
P
Sbjct: 541 P 543
>CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (12%)
Length = 312
Score = 79.3 bits (194), Expect(2) = 4e-17
Identities = 34/79 (43%), Positives = 52/79 (65%)
Frame = +1
Query: 315 LQAMGGKKPISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKR 374
L AM G+ P+SI TD D +++A+ +VFP++RHR C WHI K+ EKL+HI+ + F+
Sbjct: 1 LTAMSGRPPLSITTDHDSVIQSAIMQVFPDTRHRFCKWHIFKQCQEKLSHIFLQFPNFEA 180
Query: 375 DMKRCIRGSHSIQSFEEEW 393
+ +C+ + SI FE W
Sbjct: 181 EFHKCVNLTDSIDEFESCW 237
Score = 27.3 bits (59), Expect(2) = 4e-17
Identities = 8/23 (34%), Positives = 15/23 (64%)
Frame = +3
Query: 396 IMVEYNLKENEWLQGLYKIRESW 418
++ Y+L++NEWLQ ++ W
Sbjct: 243 LLDRYDLRDNEWLQAIHSACRQW 311
>CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sinorhizobium
meliloti}, partial (1%)
Length = 828
Score = 70.1 bits (170), Expect = 3e-12
Identities = 51/227 (22%), Positives = 91/227 (39%), Gaps = 13/227 (5%)
Frame = +1
Query: 44 IGMEFESIEKVREFYNSFDKKNGFGVRV--------RSSKPKRAVLVCCNEGQHKVKISR 95
+G S ++ + Y K GF VR K + C +G
Sbjct: 160 LGQVVNSKDEAYDLYQEHAFKTGFSVRKGKELYYDNEKKKTRLKDFYCSKQGFKN----- 324
Query: 96 TEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMR 155
+ + K + R+ C A ++ T + W + DHNH V + +R
Sbjct: 325 ----NEPDGEVAYKRADSRTNCLA-MVRFNVTKEGVWKVTKLILDHNHEFVPLQQRYLLR 489
Query: 156 CHKKMSVAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNR-----DCWNYIRNVRRKNLD 210
+ MS + ++ + I V R D NY+ + K ++
Sbjct: 490 SMRNMSNLKEDRIKSLVNDAIRVTNVGGYLGKEVGGVDKRGVMLKDMHNYVSTGKLKFIE 669
Query: 211 VGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSY 257
GDAQ++ ++ + KQ ++S F+Y++Q D +SR+ N+FW D +S + Y
Sbjct: 670 AGDAQSLLNHLQSKQAQDSMFYYSVQLDHESRLNNVFWRDGKSVVDY 810
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 58.2 bits (139), Expect = 1e-08
Identities = 28/77 (36%), Positives = 46/77 (59%)
Frame = +2
Query: 429 AGMNTTQRSESINAFFDSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILST 488
AG +T QRSES+N+FFD ++ TL+EFV ++ + +R + E ++++ HK L +
Sbjct: 2 AGTSTAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKS 181
Query: 489 GSKLEEHAGHLYTPEIF 505
S E+ +YT IF
Sbjct: 182 PSPWEKQMSTIYTHAIF 232
>TC84106 similar to GP|9502168|gb|AAF88018.1| contains simlarity to
Arabidopsis thaliana far-red impaired response protein
(GB:AAD51282.1), partial (25%)
Length = 673
Score = 39.7 bits (91), Expect(2) = 1e-07
Identities = 18/72 (25%), Positives = 36/72 (50%), Gaps = 2/72 (2%)
Frame = +2
Query: 108 RKCSTIRSGCEASLIVSRGTTK--SKWMIKSFNNDHNHVMVSPKSVSYMRCHKKMSVAAK 165
R ++R GC+A + +S+ S+W + F+N HNH ++ V + ++K+ A +
Sbjct: 395 RDRKSVRCGCDAKMYLSKDVVDGVSQWTVLQFSNVHNHELLEDDQVRLLPAYRKIHEADQ 574
Query: 166 SLVEKFEEEGIP 177
+ + G P
Sbjct: 575 ERILLLSKAGFP 610
Score = 34.7 bits (78), Expect(2) = 1e-07
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 6/86 (6%)
Frame = +1
Query: 13 DDNGIGSSSKDFTTSILDTSDIGGELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVR 72
DD+ GS++ + + SDI P +GM F+S ++ E Y +F +K+GF +R
Sbjct: 127 DDSDEGSAAPESMKADKTPSDI-----ITPYVGMVFKSDDEAFEHYGNFARKHGFSMRKA 291
Query: 73 SSK--PKRAV----LVCCNEGQHKVK 92
S+ P+ V VC G VK
Sbjct: 292 RSRISPQLGVYKRDFVCYRSGFAPVK 369
>CB893278 weakly similar to PIR|T05645|T056 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (5%)
Length = 821
Score = 53.9 bits (128), Expect = 2e-07
Identities = 34/121 (28%), Positives = 59/121 (48%), Gaps = 8/121 (6%)
Frame = +3
Query: 41 KPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKRA-------VLVCCNEG-QHKVK 92
+P IGM+F S E+ R FY+ + ++ GF VR+ ++ R VC EG + K
Sbjct: 312 EPFIGMQFNSREEARGFYDGYGRRIGFTVRIHHNRRSRVNNELIGQDFVCSKEGFRAKKY 491
Query: 93 ISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVS 152
+ R + + T R GC+A + ++ + KW++ F +H H ++SP V
Sbjct: 492 VHRKDRVLPPPPAT-------REGCQAMIRLAL-RDEGKWVVTKFVKEHTHKLMSPGEVP 647
Query: 153 Y 153
+
Sbjct: 648 W 650
>TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (66%)
Length = 1169
Score = 49.3 bits (116), Expect = 5e-06
Identities = 40/132 (30%), Positives = 58/132 (43%), Gaps = 7/132 (5%)
Frame = +2
Query: 26 TSILDTSDIGGELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRV-RSSKPKR------ 78
TS++ T D +P IG EF S + FYNS+ + GF +RV + S+ +R
Sbjct: 296 TSVMRTID-------EPYIGQEFVSEAEAHAFYNSYATRVGFVIRVSKLSRSRRDGSVIG 454
Query: 79 AVLVCCNEGQHKVKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFN 138
LVC EG + D + R+ + R GC A ++V R W I F
Sbjct: 455 RALVCNKEGFR---------MPDKREKIVRQRAETRVGCRAMIMV-RKLNSGLWSITKFV 604
Query: 139 NDHNHVMVSPKS 150
+H H + KS
Sbjct: 605 KEHTHPLTPGKS 640
>TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (7%)
Length = 354
Score = 43.1 bits (100), Expect = 3e-04
Identities = 24/54 (44%), Positives = 32/54 (58%), Gaps = 7/54 (12%)
Frame = +1
Query: 41 KPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKR---AVL----VCCNEG 87
+P GMEFES E + FYNS+ ++ GF RV SS+ R A++ VC EG
Sbjct: 190 EPSEGMEFESEEAAKAFYNSYARRVGFSTRVSSSRRSRRDGAIIQRQXVCAKEG 351
>TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (13%)
Length = 721
Score = 40.0 bits (92), Expect = 0.003
Identities = 26/102 (25%), Positives = 47/102 (45%)
Frame = +2
Query: 36 GELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKRAVLVCCNEGQHKVKISR 95
G+ + +P G+EFE+ E FY + K GF +++S+ + + K SR
Sbjct: 395 GDTDFEPPNGIEFETHEAAYSFYQEYAKSMGFTTSIKNSRRSKKTKEFIDA---KFACSR 565
Query: 96 TEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSF 137
+S ++ ++ S ++ C+A + V R KW I F
Sbjct: 566 YGVTPESDGRSSQRSSVKKTDCKACMHVKR-NPDGKWNIFEF 688
>TC89448 similar to PIR|C84864|C84864 hypothetical protein At2g43280
[imported] - Arabidopsis thaliana, partial (80%)
Length = 1177
Score = 39.7 bits (91), Expect = 0.004
Identities = 38/133 (28%), Positives = 61/133 (45%), Gaps = 6/133 (4%)
Frame = +1
Query: 11 LMDDNGIGSSSKDFTTSILDTSDIGGELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVR 70
L++ N S+K ++ L SD + +P GMEF S + + FY+ + + GF +R
Sbjct: 211 LVEPNDGCRSAKSYSGRELGISDHDNAVK-EPYEGMEFVSEDAAKIFYDEYARHVGFVMR 387
Query: 71 V----RSSKPKR--AVLVCCNEGQHKVKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVS 124
V RS + R A CN+ H V I+ ++ ++ R GC+A +I
Sbjct: 388 VMSCRRSXRDGRILARRFGCNKEGHCV------SIQGKLGPVRKPRASTREGCKA-MIHI 546
Query: 125 RGTTKSKWMIKSF 137
+ KWMI F
Sbjct: 547 KYDQSGKWMITXF 585
>TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (59%)
Length = 832
Score = 31.6 bits (70), Expect(2) = 0.005
Identities = 23/75 (30%), Positives = 32/75 (42%), Gaps = 3/75 (4%)
Frame = +2
Query: 72 RSSKPKRAV---LVCCNEGQHKVKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTT 128
RS K A+ LVC EG + D + R+ + R GC A +I+ R
Sbjct: 140 RSRKDGTAIGRALVCNKEGYR---------MPDKREKIVRQRAETRVGCRA-MIMMRKIN 289
Query: 129 KSKWMIKSFNNDHNH 143
KW+I F +H H
Sbjct: 290 SGKWVITKFVKEHTH 334
Score = 26.6 bits (57), Expect(2) = 0.005
Identities = 12/32 (37%), Positives = 15/32 (46%)
Frame = +3
Query: 36 GELNSKPEIGMEFESIEKVREFYNSFDKKNGF 67
G +P +G EFES FYNS G+
Sbjct: 15 GASAEEPYVGQEFESEAAAHAFYNSIXAARGW 110
>BF650479 homologue to PIR|T06643|T066 hypothetical protein T20K18.200 -
Arabidopsis thaliana, partial (13%)
Length = 580
Score = 38.1 bits (87), Expect = 0.011
Identities = 27/72 (37%), Positives = 39/72 (53%), Gaps = 9/72 (12%)
Frame = +2
Query: 9 HQLMDDNGIGSSSK---------DFTTSILDTSDIGGELNSKPEIGMEFESIEKVREFYN 59
HQ+ D+ + S K + +T ILD G EL +P +GMEF+S E R+FY
Sbjct: 281 HQIFMDSVVVDSDKGNEADHSFLEESTHILD----GVELQ-EPYVGMEFDSEEAARKFYA 445
Query: 60 SFDKKNGFGVRV 71
+ ++ GF VRV
Sbjct: 446 EYARRVGFVVRV 481
>BF646907 similar to GP|15983442|gb| At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (4%)
Length = 621
Score = 34.7 bits (78), Expect = 0.12
Identities = 31/105 (29%), Positives = 45/105 (42%), Gaps = 15/105 (14%)
Frame = +1
Query: 45 GMEFESIEKVREFYNSFDKKNGFGVR----VRSSKPK---------RAVLVCCNEGQHKV 91
G F S ++ FY F +KNGF +R +S K + + VC G V
Sbjct: 328 GKVFNSDDEAYNFYCLFARKNGFSIRRHHVYKSIKNQSDDNPLGVYKREFVCHRAGTISV 507
Query: 92 KISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKS--KWMI 134
KD+ + KRK + R C A L+V+ T S KW++
Sbjct: 508 D-------KDNEVEGKRKRKSSRCNCGAKLLVNITTINSXKKWVV 621
>AW696815 homologue to GP|11994736|dbj far-red impaired response protein;
Mutator-like transposase-like protein; phytochrome A
signaling, partial (7%)
Length = 394
Score = 33.1 bits (74), Expect = 0.36
Identities = 15/53 (28%), Positives = 27/53 (50%)
Frame = +2
Query: 23 DFTTSILDTSDIGGELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSK 75
D + +D + + N +P GMEFES + FY + + GF +++S+
Sbjct: 158 DLNSPTVDIAMFKEDTNLEPLSGMEFESHGEAYSFYQEYARSMGFNTAIQNSR 316
>AL386637 weakly similar to GP|13325257|gb|A Similar to CG15432 gene product
{Homo sapiens}, partial (63%)
Length = 509
Score = 32.3 bits (72), Expect = 0.61
Identities = 16/49 (32%), Positives = 25/49 (50%)
Frame = +1
Query: 456 EFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEI 504
E LKFE+ + RL+ + ++ HK R+ KLEE H P++
Sbjct: 1 EAELKFEEIQKKRLDEKVEKAARKSHKERVAELNRKLEEMTEHYDIPKV 147
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.333 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,249,956
Number of Sequences: 36976
Number of extensions: 294911
Number of successful extensions: 2330
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 1702
Number of HSP's successfully gapped in prelim test: 102
Number of HSP's that attempted gapping in prelim test: 598
Number of HSP's gapped (non-prelim): 1841
length of query: 674
length of database: 9,014,727
effective HSP length: 103
effective length of query: 571
effective length of database: 5,206,199
effective search space: 2972739629
effective search space used: 2972739629
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0253.10


