

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
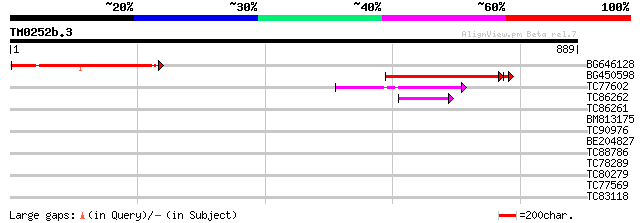
Query= TM0252b.3
(889 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG646128 weakly similar to GP|23429518|gb APC2 {Arabidopsis thal... 325 5e-89
BG450598 similar to GP|23429518|gb APC2 {Arabidopsis thaliana}, ... 296 1e-82
TC77602 similar to GP|20268719|gb|AAM14063.1 putative cullin {Ar... 59 1e-08
TC86262 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 44 2e-04
TC86261 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 42 0.001
BM813175 similar to GP|22136936|gb putative cullin 1 protein {Ar... 40 0.004
TC90976 similar to PIR|E96718|E96718 probable cullin T6C23.13 [i... 37 0.026
BE204827 similar to PIR|F86222|F86 hypothetical protein [importe... 30 3.1
TC88786 weakly similar to GP|7268619|emb|CAB78828.1 unknown prot... 30 4.1
TC78289 similar to GP|15451044|gb|AAK96793.1 Unknown protein {Ar... 30 5.3
TC80279 weakly similar to PIR|F96505|F96505 probable nucellin [i... 29 9.1
TC77569 weakly similar to GP|9757754|dbj|BAB08235.1 contains sim... 29 9.1
TC83118 similar to PIR|E96718|E96718 probable cullin T6C23.13 [i... 29 9.1
>BG646128 weakly similar to GP|23429518|gb APC2 {Arabidopsis thaliana},
partial (16%)
Length = 783
Score = 325 bits (833), Expect = 5e-89
Identities = 170/248 (68%), Positives = 198/248 (79%), Gaps = 9/248 (3%)
Frame = +2
Query: 3 LDESPSSFFNLDILNSLTQDSVHEILHSYNAFCNATQSLLAGGGGGDLSIGADFISYVNC 62
+++S SS FN+ IL++L++D +HEIL SY FCNATQSLL GG GDLS GA+F+S+V+
Sbjct: 26 MEDSHSSLFNIGILDTLSEDQLHEILESYTLFCNATQSLL--GGAGDLSYGAEFVSHVHT 199
Query: 63 LCKHGLHSLVRDHFLRVLEETFERNAAS-VWRHFEPYA--AGFSKNDDLD------DSVL 113
LCKHGL SLVRDHFL+VLEETFERN +S WRHF PYA +KN D++ +SVL
Sbjct: 200 LCKHGLESLVRDHFLKVLEETFERNGSSRFWRHFVPYADFVSLNKNGDVNIDEDEIESVL 379
Query: 114 YHVLEEICVEKHYQEKCLLILVNALQSYKDQMSEETHNFEAERNYLTSKYHWIVSSVLMA 173
+ LEEI +EK YQEKCLLILV+ALQS+KDQ SEE HNFEAERNYLTSKY W VSSVLMA
Sbjct: 380 CNALEEISLEKQYQEKCLLILVHALQSFKDQTSEERHNFEAERNYLTSKYQWTVSSVLMA 559
Query: 174 TLPPVFPVILHLYFKRRLEELSIIMDVEFHDDTSQNKDSMDLDEKGKICNNVGDMDVDDE 233
TLP VFP ILH YFKRRLEELS +MD EF DD SQNKD MDLDEKGKIC + G+MDV DE
Sbjct: 560 TLPRVFPAILHWYFKRRLEELSTVMDGEFTDDVSQNKDDMDLDEKGKICKD-GEMDV-DE 733
Query: 234 CYNNHRLS 241
CY++ R S
Sbjct: 734 CYSDRRFS 757
>BG450598 similar to GP|23429518|gb APC2 {Arabidopsis thaliana}, partial
(13%)
Length = 604
Score = 296 bits (759), Expect(2) = 1e-82
Identities = 150/186 (80%), Positives = 167/186 (89%), Gaps = 1/186 (0%)
Frame = +2
Query: 590 ISQPSQTNVEVEDNAISMDNIAATIISSNFWPPIQVEPLNLPEPVDKLLSDYAKRFNEIK 649
ISQP QT+VEVEDNAISMD +AATIISSNFWPPIQ EPLNLPEPVDKLLSDYAKRF+E+K
Sbjct: 2 ISQPPQTSVEVEDNAISMDKVAATIISSNFWPPIQDEPLNLPEPVDKLLSDYAKRFSEVK 181
Query: 650 TPRKLQWKKSLGTVKLELQLKDRVLQFTVAPVHASIIMNFQDQTSWTSKNLAAAVGIPVD 709
TPRKLQWKKSLGTVKLELQ +DR +QFTVAPV ASIIM FQDQ SWTSK+LAAA+GIPVD
Sbjct: 182 TPRKLQWKKSLGTVKLELQFEDREMQFTVAPVLASIIMKFQDQMSWTSKDLAAAIGIPVD 361
Query: 710 ALNRRMSFWISKGVVAESSGGDSSDHVYTIMESMVETKKR-DSSGITQELLGGGDEEEDR 768
LNRR++FWISKGV+AESSGGDSSDHVYTIME+M T + SG QELL GGD+EEDR
Sbjct: 362 VLNRRINFWISKGVIAESSGGDSSDHVYTIMENMAXTSRNGGGSGNAQELL-GGDDEEDR 538
Query: 769 AVASIE 774
+VAS++
Sbjct: 539 SVASVK 556
Score = 29.6 bits (65), Expect(2) = 1e-82
Identities = 12/16 (75%), Positives = 14/16 (87%)
Frame = +1
Query: 775 NQLRKEMSIYEKFIMG 790
NQLRKEM+ YEKF +G
Sbjct: 556 NQLRKEMTXYEKFXLG 603
>TC77602 similar to GP|20268719|gb|AAM14063.1 putative cullin {Arabidopsis
thaliana}, partial (87%)
Length = 2708
Score = 58.5 bits (140), Expect = 1e-08
Identities = 49/206 (23%), Positives = 88/206 (41%), Gaps = 1/206 (0%)
Frame = +2
Query: 511 VDILGMIVGIIGSKDQLVHEYRTMLAEKLLNKSDYDIDSEIRTLELLKIHFGESSLQKCE 570
+D + ++ I KD Y+ LA++LL ID+E + LK G K E
Sbjct: 1133 LDKVLVLFRFIQGKDVFEAFYKKDLAKRLLLGKSASIDAEKSMISKLKTECGSQFTNKLE 1312
Query: 571 IMLNDLIGSKRINSNIKATISQPSQTNVEVEDNAISMDNIAATIISSNFWPPIQVEPLNL 630
M D+ SK IN + + Q SQ ++ ++ ++++ +WP + L
Sbjct: 1313 GMFKDIELSKEINESFR----QSSQARTKLPSGI----EMSVHVLTTGYWPTYPPMDVRL 1468
Query: 631 PEPVDKLLSDYAKRFNEIK-TPRKLQWKKSLGTVKLELQLKDRVLQFTVAPVHASIIMNF 689
P ++ + D K F K + R+L W+ SLG L+ + V+ ++M F
Sbjct: 1469 PHELN-VYQDIFKEFYLSKYSGRRLMWQNSLGHCVLKADFPKGKKELAVSLFQTVVLMQF 1645
Query: 690 QDQTSWTSKNLAAAVGIPVDALNRRM 715
D + +++ + GI L R +
Sbjct: 1646 NDAEKLSFQDIKDSTGIEDKELRRTL 1723
>TC86262 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, complete
Length = 2332
Score = 44.3 bits (103), Expect = 2e-04
Identities = 23/89 (25%), Positives = 45/89 (49%), Gaps = 2/89 (2%)
Frame = +2
Query: 610 IAATIISSNFWPPIQVEPLNLPEPVDKLLSDYAKRFNEIKTPRKLQWKKSLGTVKLELQL 669
+ T++++ FWP + LNLP + K + + + ++ RKL W SLGT + +
Sbjct: 1646 LTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF 1825
Query: 670 KDRVLQFTVAPVHAS--IIMNFQDQTSWT 696
+ ++ V AS ++ N D+ S++
Sbjct: 1826 DPKTVELVVTTYQASALLLFNSSDRLSYS 1912
>TC86261 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (72%)
Length = 1742
Score = 42.0 bits (97), Expect = 0.001
Identities = 30/120 (25%), Positives = 50/120 (41%)
Frame = +3
Query: 517 IVGIIGSKDQLVHEYRTMLAEKLLNKSDYDIDSEIRTLELLKIHFGESSLQKCEIMLNDL 576
++ I KD YR LA +LL + D E L LK G K E M+ DL
Sbjct: 1386 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 1565
Query: 577 IGSKRINSNIKATISQPSQTNVEVEDNAISMDNIAATIISSNFWPPIQVEPLNLPEPVDK 636
+K ++ + + + ++ + T++++ FWP + LNLP + K
Sbjct: 1566 TLAKENQTSFEEYLKNNPNVDPGID--------LTVTVLTTGFWPSYKSFDLNLPAEMVK 1721
>BM813175 similar to GP|22136936|gb putative cullin 1 protein {Arabidopsis
thaliana}, partial (30%)
Length = 680
Score = 40.0 bits (92), Expect = 0.004
Identities = 26/87 (29%), Positives = 47/87 (53%), Gaps = 3/87 (3%)
Frame = +1
Query: 613 TIISSNFWPPIQVEPLNLPEPVDKLLSDYAKRFNEIKTP-RKLQWKKSLGTVKLELQLKD 671
T++++ WP + LNLP + K + + K F E KT RKL W SLGT + + +
Sbjct: 4 TVLTTVLWPSYKSFDLNLPSEMVKCVEVF-KGFYETKTKHRKLTWIYSLGTCNIIGKFEP 180
Query: 672 RVLQFTVAPVHAS--IIMNFQDQTSWT 696
+ ++ V+ A+ ++ N D+ S++
Sbjct: 181 KTIELIVSTYQAAALLLFNTADKLSYS 261
>TC90976 similar to PIR|E96718|E96718 probable cullin T6C23.13 [imported] -
Arabidopsis thaliana, partial (33%)
Length = 825
Score = 37.4 bits (85), Expect = 0.026
Identities = 22/105 (20%), Positives = 43/105 (40%), Gaps = 1/105 (0%)
Frame = +3
Query: 610 IAATIISSNFWPPIQVEPLNLPEPVDKLLSDYAKRFNEIKTPRKLQWKKSLGTVKLELQL 669
+ ++++ WP NLP + L + + T R+L W+ ++G L+
Sbjct: 6 LTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATF 185
Query: 670 -KDRVLQFTVAPVHASIIMNFQDQTSWTSKNLAAAVGIPVDALNR 713
K + + V+ ++M F + + K + A IP L R
Sbjct: 186 GKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKR 320
>BE204827 similar to PIR|F86222|F86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (18%)
Length = 626
Score = 30.4 bits (67), Expect = 3.1
Identities = 29/119 (24%), Positives = 42/119 (34%), Gaps = 7/119 (5%)
Frame = -3
Query: 57 ISYVNCLCKHGLHSLVRDHFLRVLEETFERNAASVWRHFEPYAAGFS-------KNDDLD 109
+SY+NC LHS + LE+ F+ +A S + G S K
Sbjct: 354 VSYINCSTSARLHSTLS------LEDLFQHHATSFQVQLQNIVLGGSALPREALKRATRA 193
Query: 110 DSVLYHVLEEICVEKHYQEKCLLILVNALQSYKDQMSEETHNFEAERNYLTSKYHWIVS 168
++V Y V E + N + +S E H E L YH I+S
Sbjct: 192 NTVTYPVQVSHVSEPLLNYHSVSKFYNCIYHDMQDISSEFHPMECRNTMLICSYHSILS 16
>TC88786 weakly similar to GP|7268619|emb|CAB78828.1 unknown protein
{Arabidopsis thaliana}, partial (22%)
Length = 1166
Score = 30.0 bits (66), Expect = 4.1
Identities = 17/44 (38%), Positives = 24/44 (53%), Gaps = 3/44 (6%)
Frame = +1
Query: 292 RGSILQSI---QRWIQAVPLQFLHALLVYIGDSVSYESTSSGLK 332
RGS +S+ WI + FL L VYIG + +E TS G++
Sbjct: 718 RGSKRRSVWFFAHWILGTAVTFLGVLNVYIGLAAYHEKTSKGIR 849
>TC78289 similar to GP|15451044|gb|AAK96793.1 Unknown protein {Arabidopsis
thaliana}, partial (15%)
Length = 1020
Score = 29.6 bits (65), Expect = 5.3
Identities = 21/60 (35%), Positives = 27/60 (45%), Gaps = 4/60 (6%)
Frame = -3
Query: 53 GADFISYVNCLCKHGLHSLVRDH----FLRVLEETFERNAASVWRHFEPYAAGFSKNDDL 108
G+ + Y+ CL H L S H FL F R A V P+ GFS+NDD+
Sbjct: 475 GSAAVKYL-CLVFHPLSSNKNQHPGALFLAAPHIWFSRPIAQVLGFLTPHHQGFSQNDDV 299
>TC80279 weakly similar to PIR|F96505|F96505 probable nucellin [imported] -
Arabidopsis thaliana, partial (42%)
Length = 835
Score = 28.9 bits (63), Expect = 9.1
Identities = 22/58 (37%), Positives = 27/58 (45%), Gaps = 1/58 (1%)
Frame = +3
Query: 7 PSSFFNLDILNSLTQDSVHEILHSYNAFCNATQSLLAGGGGGDLSIGADFISYV-NCL 63
PS+ L + N T S+ LHS N L+G GGG L G D I + NCL
Sbjct: 666 PSTTGVLGLGNGRT--SILSQLHSLGLIRNVVGHCLSGRGGGFLFFGDDLIPLIRNCL 833
>TC77569 weakly similar to GP|9757754|dbj|BAB08235.1 contains similarity to
root cap protein~gene_id:MUF9.15 {Arabidopsis thaliana},
partial (26%)
Length = 835
Score = 28.9 bits (63), Expect = 9.1
Identities = 27/102 (26%), Positives = 44/102 (42%), Gaps = 7/102 (6%)
Frame = -1
Query: 594 SQTNVEVEDNAISMDNIAATIISSNFWPPIQVEPLNLP---EPVDKLLSDYAKRFNEIKT 650
S ++ +++D S++ + + + N IQV L LP P+ + L ++K
Sbjct: 835 STSSSQIDDGVASIEKLCESNRTPN--A*IQV*SLGLPCGLRPIKRALI*RLVSDTKLKC 662
Query: 651 PRKLQWK----KSLGTVKLELQLKDRVLQFTVAPVHASIIMN 688
L WK S T + R LQF + P+H IMN
Sbjct: 661 SFLLPWK*KTMPSAPTNRGSKHADPRPLQFALRPIHIQYIMN 536
>TC83118 similar to PIR|E96718|E96718 probable cullin T6C23.13 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 902
Score = 28.9 bits (63), Expect = 9.1
Identities = 12/57 (21%), Positives = 25/57 (43%)
Frame = +3
Query: 610 IAATIISSNFWPPIQVEPLNLPEPVDKLLSDYAKRFNEIKTPRKLQWKKSLGTVKLE 666
+ ++++ WP NLP + L + + T R+L W+ ++G L+
Sbjct: 600 LTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLK 770
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,311,628
Number of Sequences: 36976
Number of extensions: 358845
Number of successful extensions: 1959
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1953
length of query: 889
length of database: 9,014,727
effective HSP length: 105
effective length of query: 784
effective length of database: 5,132,247
effective search space: 4023681648
effective search space used: 4023681648
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0252b.3


