

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
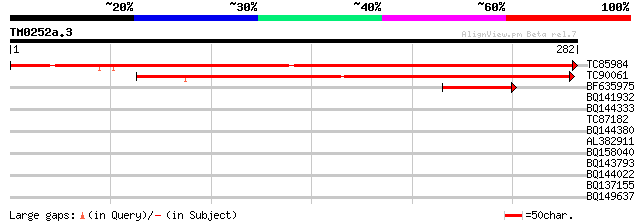
Query= TM0252a.3
(282 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85984 homologue to GP|15076863|gb|AAK82969.1 putative heme oxy... 427 e-120
TC90061 similar to PIR|H84661|H84661 heme oxygenase 2 (HO2) [imp... 230 4e-61
BF635975 similar to GP|15076863|gb| putative heme oxygenase 1 pr... 68 3e-12
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 29 2.3
BQ144333 weakly similar to GP|7331719|gb|A Hypothetical protein ... 26 2.7
TC87182 similar to GP|18176296|gb|AAL60018.1 unknown protein {Ar... 28 3.0
BQ144380 similar to GP|1185397|gb|A SH3 domain binding protein {... 28 5.2
AL382911 similar to GP|21902044|dbj hypothetical protein~similar... 28 5.2
BQ158040 28 5.2
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 28 5.2
BQ144022 homologue to GP|22831315|dbj hypothetical protein~predi... 27 6.8
BQ137155 27 8.8
BQ149637 similar to PIR|D85135|D85 hypothetical protein AT4g1261... 27 8.8
>TC85984 homologue to GP|15076863|gb|AAK82969.1 putative heme oxygenase 1
precursor {Pisum sativum}, partial (84%)
Length = 1412
Score = 427 bits (1097), Expect = e-120
Identities = 220/285 (77%), Positives = 245/285 (85%), Gaps = 3/285 (1%)
Frame = +2
Query: 1 MASATVVSQIQSLYIIKPRLSPPPPPHRQFRSIYFPTTRLLQQ--HRFRQMK-SVVIVSA 57
MAS T + QIQS++ K P QFRS +F +L +R MK S VIVSA
Sbjct: 401 MASLTPLYQIQSIFHYKTNY--PQFSTHQFRSNFFFQKKLSFSSFNRMPPMKQSTVIVSA 574
Query: 58 TAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYL 117
T+A +KR+PGESKGFVEEMRFVAM+LHTKDQAKEGEKEVTEPEE+AVTKWEP+VDGYL
Sbjct: 575 TSAAAEKKRHPGESKGFVEEMRFVAMRLHTKDQAKEGEKEVTEPEERAVTKWEPSVDGYL 754
Query: 118 KFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPG 177
+FLVDSK+VYDTLEKIVQDA + YAEF+NTGLERSA+L KDL+WFKEQGYTIP+PSSPG
Sbjct: 755 RFLVDSKIVYDTLEKIVQDAAY--YAEFKNTGLERSANLDKDLKWFKEQGYTIPEPSSPG 928
Query: 178 LNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLS 237
L YAQYL DLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAG+LLN++ LEFYKWDGDL
Sbjct: 929 LTYAQYLTDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGQLLNDQALEFYKWDGDLK 1108
Query: 238 QLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
QLLQNVRDKLNKVAE+WTREEK+HCLEETEKSFK+SGEILRLILS
Sbjct: 1109QLLQNVRDKLNKVAEEWTREEKDHCLEETEKSFKFSGEILRLILS 1243
>TC90061 similar to PIR|H84661|H84661 heme oxygenase 2 (HO2) [imported] -
Arabidopsis thaliana, partial (47%)
Length = 999
Score = 230 bits (587), Expect = 4e-61
Identities = 113/221 (51%), Positives = 155/221 (70%), Gaps = 3/221 (1%)
Frame = +3
Query: 64 RKRYPGESKGFVEEMRFVAMKLH---TKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFL 120
RK YPGE+ G EEMRFVAMKLH T V + E + W P++ G+L+FL
Sbjct: 240 RKLYPGETIGITEEMRFVAMKLHNDKTNTVNNTTSVVVEDAEGQIPDTWYPSMKGFLRFL 419
Query: 121 VDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNY 180
VD+++V+ TLE+IV D+ + SYA R TGLERS + KDLEW KE G IP PSSPG Y
Sbjct: 420 VDNQLVFSTLERIVDDSDNVSYAYLRKTGLERSEGILKDLEWLKE-GVEIPNPSSPGTTY 596
Query: 181 AQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLL 240
A+YL +L++ F+ HFYNI+F+H G++I KK++ +LL K LEF KW+GD+ ++L
Sbjct: 597 AKYLEELAERSAPLFLSHFYNIHFSHITAGQVITKKVSEKLLEGKELEFCKWEGDVQEML 776
Query: 241 QNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLIL 281
++VR+KLN +AE W R+EKN CL ET+KSF++ G+I+RLI+
Sbjct: 777 KDVREKLNVLAEHWGRDEKNKCLRETKKSFQFMGQIVRLII 899
>BF635975 similar to GP|15076863|gb| putative heme oxygenase 1 precursor
{Pisum sativum}, partial (12%)
Length = 655
Score = 68.2 bits (165), Expect = 3e-12
Identities = 31/37 (83%), Positives = 35/37 (93%)
Frame = +1
Query: 216 KIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAE 252
+IAG+LLN++ LEFYKWDGDL QLLQNVRDKLNKVAE
Sbjct: 469 QIAGQLLNDQALEFYKWDGDLKQLLQNVRDKLNKVAE 579
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 28.9 bits (63), Expect = 2.3
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 3/42 (7%)
Frame = -3
Query: 2 ASATVVSQIQSLYIIK---PRLSPPPPPHRQFRSIYFPTTRL 40
A++ V ++ LY ++ PR PPPPPH SI P++RL
Sbjct: 373 AASAVPLPLRPLYPLRDSPPRRPPPPPPHSP--SILPPSSRL 254
>BQ144333 weakly similar to GP|7331719|gb|A Hypothetical protein Y110A2AL.12
{Caenorhabditis elegans}, partial (8%)
Length = 1163
Score = 26.2 bits (56), Expect(2) = 2.7
Identities = 10/30 (33%), Positives = 19/30 (63%)
Frame = -1
Query: 54 IVSATAAETARKRYPGESKGFVEEMRFVAM 83
I++ + +K++PGE+KGF + F A+
Sbjct: 599 ILAGAKKKKKKKKFPGETKGFFFFLTFFAL 510
Score = 20.8 bits (42), Expect(2) = 2.7
Identities = 9/19 (47%), Positives = 10/19 (52%)
Frame = -3
Query: 12 SLYIIKPRLSPPPPPHRQF 30
SLY I+ R PP R F
Sbjct: 774 SLYYIRARAGPPSKKKRGF 718
>TC87182 similar to GP|18176296|gb|AAL60018.1 unknown protein {Arabidopsis
thaliana}, complete
Length = 2064
Score = 28.5 bits (62), Expect = 3.0
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Frame = +1
Query: 59 AAETARKRYPGESKG--FVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGY 116
AAE+ + G +KG + ++ V +L+ ++ + +E+ EKA T W TV GY
Sbjct: 949 AAESLHQEERGGAKGRKHRKNVKAVEKELYQLEEDVKLLEEMYPQGEKAETSWALTVLGY 1128
Query: 117 LKFLV 121
L LV
Sbjct: 1129 LAKLV 1143
>BQ144380 similar to GP|1185397|gb|A SH3 domain binding protein {Rattus
norvegicus}, partial (12%)
Length = 1269
Score = 27.7 bits (60), Expect = 5.2
Identities = 11/27 (40%), Positives = 13/27 (47%)
Frame = -1
Query: 18 PRLSPPPPPHRQFRSIYFPTTRLLQQH 44
P PPPPPHR + + P Q H
Sbjct: 210 PTPHPPPPPHRPRMTTHSPRRNQAQHH 130
>AL382911 similar to GP|21902044|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 At5g25830, partial
(6%)
Length = 261
Score = 27.7 bits (60), Expect = 5.2
Identities = 13/40 (32%), Positives = 20/40 (49%)
Frame = +3
Query: 14 YIIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVV 53
+ ++PR PPPPPH + + LLQ+ M+ V
Sbjct: 138 FSLQPRPPPPPPPHLLTLTPWLHQKSLLQERETPMMEGRV 257
>BQ158040
Length = 1017
Score = 27.7 bits (60), Expect = 5.2
Identities = 14/36 (38%), Positives = 21/36 (57%), Gaps = 6/36 (16%)
Frame = +2
Query: 20 LSPPPPPHRQFR------SIYFPTTRLLQQHRFRQM 49
L+PPP PHR + SI+ PT + QHR +++
Sbjct: 311 LTPPPRPHRCWEIVFVDTSIFIPTQQTNIQHRQKRV 418
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 27.7 bits (60), Expect = 5.2
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +1
Query: 14 YIIKPRLSPPPPPH 27
++ K LSPPPPPH
Sbjct: 169 FLFKDHLSPPPPPH 210
>BQ144022 homologue to GP|22831315|dbj hypothetical protein~predicted by
GlimmerM etc. {Oryza sativa (japonica cultivar-group)},
partial (16%)
Length = 1286
Score = 27.3 bits (59), Expect = 6.8
Identities = 20/51 (39%), Positives = 25/51 (48%), Gaps = 8/51 (15%)
Frame = -3
Query: 18 PRLSPPPPPHRQF-----RSIYFPTTRLL---QQHRFRQMKSVVIVSATAA 60
PR PPP P R RS P RLL + R R++ S V+ SA A+
Sbjct: 915 PRARPPPAPSRLLPLPTPRSRCLPRPRLLVRVRARRSRRVHSSVVPSARAS 763
>BQ137155
Length = 1005
Score = 26.9 bits (58), Expect = 8.8
Identities = 10/19 (52%), Positives = 13/19 (67%)
Frame = -2
Query: 23 PPPPHRQFRSIYFPTTRLL 41
PPP RQF I+ P+ R+L
Sbjct: 227 PPPSQRQFPHIHIPSARIL 171
>BQ149637 similar to PIR|D85135|D85 hypothetical protein AT4g12610 [imported]
- Arabidopsis thaliana, partial (15%)
Length = 538
Score = 26.9 bits (58), Expect = 8.8
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = -2
Query: 2 ASATVVSQIQSLYIIKPRLSPPPPPHR 28
A+++ +SQI S + PPPPPHR
Sbjct: 342 AASSALSQIYSDLPLLHYXHPPPPPHR 262
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,651,903
Number of Sequences: 36976
Number of extensions: 116798
Number of successful extensions: 1346
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1216
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1332
length of query: 282
length of database: 9,014,727
effective HSP length: 95
effective length of query: 187
effective length of database: 5,502,007
effective search space: 1028875309
effective search space used: 1028875309
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0252a.3


