

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0250.10
(1065 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
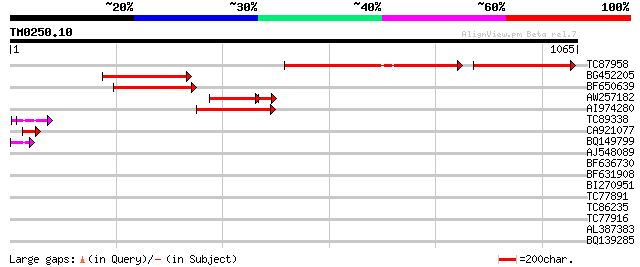
Score E
Sequences producing significant alignments: (bits) Value
TC87958 weakly similar to GP|20279447|gb|AAM18727.1 putative TNP... 278 e-132
BG452205 weakly similar to GP|1345502|emb TNP2 {Antirrhinum maju... 249 4e-66
BF650639 weakly similar to GP|22748425|g putative transposon pro... 225 6e-59
AW257182 weakly similar to GP|20279447|g putative TNP2-like tran... 167 2e-49
AI974280 weakly similar to GP|20279447|g putative TNP2-like tran... 193 3e-49
TC89338 56 6e-08
CA921077 51 3e-06
BQ149799 45 1e-04
AJ548089 41 0.003
BF636730 39 0.011
BF631908 weakly similar to GP|21450441|gb| putative transposon p... 35 0.12
BI270951 homologue to GP|20906098|gb Cobalamin biosynthesis prot... 33 0.58
TC77891 weakly similar to SP|P93147|C81E_GLYEC Cytochrome P450 8... 31 2.2
TC86235 30 6.5
TC77916 similar to GP|17979073|gb|AAL49804.1 putative gamma glut... 30 6.5
AL387383 similar to GP|6682235|gb| unknown protein {Arabidopsis ... 29 8.4
BQ139285 weakly similar to PIR|T05099|T05 hypothetical protein F... 29 8.4
>TC87958 weakly similar to GP|20279447|gb|AAM18727.1 putative TNP2-like
transposon protein {Oryza sativa (japonica
cultivar-group)}, partial (10%)
Length = 2433
Score = 278 bits (712), Expect(2) = e-132
Identities = 181/340 (53%), Positives = 221/340 (64%), Gaps = 7/340 (2%)
Frame = +2
Query: 517 NCLIGPIIHYVITLM*CTSKRTFVITLLELC*ISQGRQKTILMLVMICKTWALEKIFILG 576
+C IG IHY+I LM*C KR F+IT L L I Q RQK +L +VMI K WALEKIFI G
Sbjct: 2 SCHIGLAIHYIIILM*CILKRIFLITSLGLYWIYQERQKIMLEVVMIWKKWALEKIFIQG 181
Query: 577 MLVEVVQSL*KHVFQ*LQKRSLLFVVF*RLPNYQMALHPIFQDV*MSLRRKYLVIRVTMH 636
MLV VV L HV *L KRSL FVVF*R+ NYQM +H IFQDV* L+RKY+VIRV MH
Sbjct: 182 MLVGVVSRLQIHVSP*LLKRSLCFVVF*RVQNYQMVVHQIFQDV*KFLKRKYVVIRVMMH 361
Query: 637 ISCCITYYR*Q*EAQCLMECSHLSFVLVLFSVLCAKKLSKCKI*ITYKQRLWKYFVNWR* 696
ISCCIT R Q EAQCL + +LSFVLV+FSVL AKK+ + ++ + + + R
Sbjct: 362 ISCCITCCRYQYEAQCLKQWPNLSFVLVVFSVLYAKKVIQIHDLDNLQEEIAEVLCSTRD 541
Query: 697 FFLLVSLT*WFTC---LSISQMR---LDWVVRFSSDGCIPLKDIYVSLRVMFVIEIIQKV 750
F +*+F C LS S + W + DGCIP KDIYVSL+ MFVIE++ KV
Sbjct: 542 GF----SS*FF*CYGSLSYSSFE*G*IGW-PSSTFDGCIP*KDIYVSLKDMFVIEVVPKV 706
Query: 751 QLLRGIWLKKH*LFAQDTCIKVYRQG*IERVETTITMVHVK*MKLITFQLLVVH*G-GKM 809
LLR +WLK+ *LF QD C KV+ QG ER+ T + +++ +*MKLITFQ+LVV *G KM
Sbjct: 707 LLLRVMWLKRA*LFVQDICTKVWIQGQTERIRTMMIVIY*R*MKLITFQVLVVL*GERKM 886
Query: 810 GNRLPWI*SLWPKRIDISYSIVIVFKFTSGLQSYPCLPTL 849
G+ WI P+ ID SYSIV K+ S + TL
Sbjct: 887 GSHFIWILIH*PRPIDTSYSIVEKLKYISESMKVLLIATL 1006
Score = 211 bits (538), Expect(2) = e-132
Identities = 123/192 (64%), Positives = 141/192 (73%)
Frame = +3
Query: 872 YC*QSH*KTKVG*SQGSKSRF**VV*NSSRGR*CTIPTSRIF*RTKYCGKEVLRLPH*WI 931
YC* H*KT VG*SQGS F**+V NSS+ R*CT RI *RTK C K+V R+ *WI
Sbjct: 987 YC**PH*KT*VG*SQGSMPEF**LVQNSSQER*CTTSAKRIL*RTK*CCKKVFRISC*WI 1166
Query: 932 *IPYCTTRCETQNSEFWCYFGFVDFKFC*L*G*KSQD*TCNLLWSH**YNRVRLLWALQI 991
*IPY RC+TQ+S+FWC FGFV+ KFC L *KS+D T LWS+**YN V LLW LQI
Sbjct: 1167 *IPYYAMRCKTQDSKFWCDFGFVNCKFCKLER*KSKDRTGQFLWSN**YN*VELLWPLQI 1346
Query: 992 CLISV*LV*S*RRQIWTYLCTLQQKMLSK*SFCATYSSPPVFLC*RSFERK*VLCYENSS 1051
C I V*LV*S*RR IWTYLC LQ K+LS+* FC +SS +FLC RSFE K* LC+E+ S
Sbjct: 1347 CSI*V*LV*S*RR*IWTYLCVLQ*KVLSR*CFCDCFSSSAMFLCSRSFECK*TLCFEDRS 1526
Query: 1052 TRIIQHG*SIRV 1063
R + +G* IRV
Sbjct: 1527 KRFV*NG*PIRV 1562
>BG452205 weakly similar to GP|1345502|emb TNP2 {Antirrhinum majus}, partial
(16%)
Length = 619
Score = 249 bits (636), Expect = 4e-66
Identities = 108/167 (64%), Positives = 135/167 (80%)
Frame = +1
Query: 174 LNIPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKEFPQVAG 233
L +P+S+NKT+ MIRDLGLDY +I ACPNDCMLYWK+++ND SC C ASRW E +G
Sbjct: 115 LKVPDSYNKTR*MIRDLGLDYSKIDACPNDCMLYWKDHKNDISCHVCGASRWIESTTESG 294
Query: 234 EFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQAW 293
+ E+++ E+KV K+LRHFPLIPRLQRLFMCS T+ S+RWH ++ SKDGKLRHP DGQAW
Sbjct: 295 QIEENKKEHKVSTKILRHFPLIPRLQRLFMCSETATSLRWHHDDCSKDGKLRHPADGQAW 474
Query: 294 QDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYN 340
+DF+G H +FA D RN+RLGLASDGFNPFR M++ + WPV L+ YN
Sbjct: 475 KDFNGHHPEFARDSRNIRLGLASDGFNPFRAMNLKYXAWPVXLIPYN 615
>BF650639 weakly similar to GP|22748425|g putative transposon protein {Oryza
sativa (japonica cultivar-group)}, partial (6%)
Length = 568
Score = 225 bits (574), Expect = 6e-59
Identities = 96/156 (61%), Positives = 121/156 (77%)
Frame = +2
Query: 196 RIHACPNDCMLYWKEYENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLI 255
+I AC NDCMLYWK+++ND SC C ASR E G+ E+++ E+KV K+LRHFPLI
Sbjct: 71 KIDACXNDCMLYWKDHKNDISCHVCGASRXIESTTEXGQIEENKKEHKVSTKILRHFPLI 250
Query: 256 PRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLA 315
P LQR FMCS T+ S+RWH ++ SKDGKLRHP DGQAW+DF+G H +FA D RN+ LGLA
Sbjct: 251 PXLQRXFMCSETATSLRWHHDDXSKDGKLRHPADGQAWKDFNGHHPEFARDSRNIXLGLA 430
Query: 316 SDGFNPFRTMSISHSTWPVLLVVYNYPPWLCMKSEY 351
SDGFNPFR M++ +S WP +L+ YN+PPWLCMK+ Y
Sbjct: 431 SDGFNPFRAMNLKYSAWPXILIPYNFPPWLCMKAXY 538
>AW257182 weakly similar to GP|20279447|g putative TNP2-like transposon
protein {Oryza sativa (japonica cultivar-group)},
partial (6%)
Length = 387
Score = 167 bits (424), Expect(2) = 2e-49
Identities = 75/96 (78%), Positives = 80/96 (83%)
Frame = +1
Query: 376 IEELKELWEVGVETYDASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNT 435
IEELK LWE GVETYDAS+ + F+M AALLWTI+D+PAYAMLSGWSTKGKLAC CCN NT
Sbjct: 7 IEELKVLWESGVETYDASKNQMFRMRAALLWTISDFPAYAMLSGWSTKGKLACPCCNNNT 186
Query: 436 SSTYLKHSHKMCYMDHRVFLPADHTWRTTENLFNGK 471
SSTYLKHS KMCYMDHR FLP DH WR FNGK
Sbjct: 187 SSTYLKHSRKMCYMDHRTFLPMDHAWRENRKSFNGK 294
Score = 48.1 bits (113), Expect(2) = 2e-49
Identities = 23/34 (67%), Positives = 28/34 (81%)
Frame = +3
Query: 467 LFNGKMEFRTASPLLKGTEILEILKNFNNVFGKT 500
+F K E R+A +L+GTEILEILK+FNNVFGKT
Sbjct: 279 IF*WKKELRSAPTMLEGTEILEILKDFNNVFGKT 380
>AI974280 weakly similar to GP|20279447|g putative TNP2-like transposon
protein {Oryza sativa (japonica cultivar-group)},
partial (8%)
Length = 457
Score = 193 bits (490), Expect = 3e-49
Identities = 93/149 (62%), Positives = 107/149 (71%)
Frame = +1
Query: 351 YMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWTIND 410
+ M+SLLIPG SP +NIDVYL+PLI+ELKELWE G E YDAS K+ F+ HAALLWTI+D
Sbjct: 7 HQMLSLLIPGPSSPKRNIDVYLEPLIDELKELWERGFEVYDASSKQLFEAHAALLWTISD 186
Query: 411 YPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMDHRVFLPADHTWRTTENLFNG 470
PAY MLSGWST GKLAC C+Y+T S YLKHS K CYM HR FL H WR + F+G
Sbjct: 187 LPAYTMLSGWSTSGKLACPVCSYDTCSMYLKHSRKTCYMSHRRFLDPKHRWRKDKKGFDG 366
Query: 471 KMEFRTASPLLKGTEILEILKNFNNVFGK 499
K E R A L G+ IL L N N+FGK
Sbjct: 367 KKEIRVAPFPLSGSAILRKLGNRQNLFGK 453
>TC89338
Length = 1088
Score = 56.2 bits (134), Expect = 6e-08
Identities = 33/79 (41%), Positives = 40/79 (49%), Gaps = 2/79 (2%)
Frame = +3
Query: 3 KEWTKQPRFSSEYINGVESFLDFAYTKGRPQGDEIL-CPCAKCGNCCWE-KRNVIYNHLI 60
K W K R S+EY NGV FL FA K P + + CPC CGN + + I HL+
Sbjct: 222 KRWMKAYRLSAEYDNGVTEFLQFA-EKNLPNTEGLFPCPCVSCGNRDPKLSKEEIKGHLV 398
Query: 61 CTGFLEGYNVWIHHGEQMM 79
G + Y I HGE MM
Sbjct: 399 SVGICQNYTQ*IWHGETMM 455
Score = 53.1 bits (126), Expect = 5e-07
Identities = 29/70 (41%), Positives = 36/70 (51%), Gaps = 4/70 (5%)
Frame = +3
Query: 14 EYINGVESFLDFAYTKGRPQGDEIL-CPCAKCGNCCWE---KRNVIYNHLICTGFLEGYN 69
EY NG+ FL FA K P + + CPC CGN W+ + I HL+ G + Y
Sbjct: 498 EYDNGMTEFLQFA-EKNLPNTEGLFPCPCISCGN--WDPKLSKEEIRGHLVSVGICQNYT 668
Query: 70 VWIHHGEQMM 79
WI HGE MM
Sbjct: 669 EWIWHGETMM 698
>CA921077
Length = 438
Score = 50.8 bits (120), Expect = 3e-06
Identities = 20/35 (57%), Positives = 26/35 (74%)
Frame = -2
Query: 24 DFAYTKGRPQGDEILCPCAKCGNCCWEKRNVIYNH 58
D A+TKG + +EILCPCA C N WE R+V+Y+H
Sbjct: 437 DIAFTKGMVEEEEILCPCAVCCNDSWETRDVVYDH 333
>BQ149799
Length = 437
Score = 45.4 bits (106), Expect = 1e-04
Identities = 23/47 (48%), Positives = 26/47 (54%), Gaps = 1/47 (2%)
Frame = +1
Query: 1 MDKEWTKQPRFSSEYINGVESFLDFAYTKGRPQGDEIL-CPCAKCGN 46
MD+ W K R S EY NGV FL FA K P + + CPC CGN
Sbjct: 259 MDRRWMKAYRLSVEYDNGVTEFLQFA-EKNLPNTEGLFPCPCVSCGN 396
>AJ548089
Length = 560
Score = 40.8 bits (94), Expect = 0.003
Identities = 18/44 (40%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Frame = +1
Query: 39 CPCAKCGNCCWE---KRNVIYNHLICTGFLEGYNVWIHHGEQMM 79
CPC CGN W+ + I HL+ G + Y WI +GE MM
Sbjct: 241 CPCISCGN--WDPKLSKEEIRGHLVSVGICQNYTEWIWYGETMM 366
>BF636730
Length = 637
Score = 38.9 bits (89), Expect = 0.011
Identities = 24/71 (33%), Positives = 36/71 (49%), Gaps = 3/71 (4%)
Frame = +1
Query: 10 RFSSEYINGVESFLDFAYTKGR-PQGDEILCPCAKCGNCCWEKRNV--IYNHLICTGFLE 66
R + E++ GV F++FA + + ++ CPC C C + +V + HL GF E
Sbjct: 340 RVTDEFLAGVCEFVEFACAQDDFKKVGKLRCPCKICK--CRKP*SVDDVMKHLCMKGFKE 513
Query: 67 GYNVWIHHGEQ 77
Y W HGEQ
Sbjct: 514 DYYYWSSHGEQ 546
>BF631908 weakly similar to GP|21450441|gb| putative transposon protein
5'-partial {Oryza sativa (japonica cultivar-group)},
partial (17%)
Length = 681
Score = 35.4 bits (80), Expect = 0.12
Identities = 20/61 (32%), Positives = 36/61 (58%)
Frame = +1
Query: 715 MRLDWVVRFSSDGCIPLKDIYVSLRVMFVIEIIQKVQLLRGIWLKKH*LFAQDTCIKVYR 774
M+L ++ + GCI L+ +++ R+M+ IE IQ+VQL + + * Q T + V++
Sbjct: 433 MKLGLPDQYITVGCIQLRGLFLH*RIMYEIETIQRVQLQKDM*QMNV*HTVQGT*VMVFK 612
Query: 775 Q 775
Q
Sbjct: 613 Q 615
>BI270951 homologue to GP|20906098|gb Cobalamin biosynthesis protein
{Methanosarcina mazei Goe1}, partial (0%)
Length = 432
Score = 33.1 bits (74), Expect = 0.58
Identities = 14/28 (50%), Positives = 17/28 (60%)
Frame = +1
Query: 3 KEWTKQPRFSSEYINGVESFLDFAYTKG 30
K+W P S Y +GV FLD A+TKG
Sbjct: 337 KDWMALPPHSQSYKDGVNYFLDIAFTKG 420
>TC77891 weakly similar to SP|P93147|C81E_GLYEC Cytochrome P450 81E1 (EC
1.14.-.-) (Isoflavone 2'-hydroxylase) (P450 91A4) (CYP
GE-3). [Licorice], partial (91%)
Length = 1707
Score = 31.2 bits (69), Expect = 2.2
Identities = 10/22 (45%), Positives = 18/22 (81%)
Frame = -1
Query: 337 VVYNYPPWLCMKSEYMMMSLLI 358
V+YN WLC+K +++M+S++I
Sbjct: 1707 VIYNLSSWLCIKRKHVMLSMII 1642
>TC86235
Length = 2393
Score = 29.6 bits (65), Expect = 6.5
Identities = 14/46 (30%), Positives = 30/46 (64%)
Frame = +3
Query: 540 VITLLELC*ISQGRQKTILMLVMICKTWALEKIFILGMLVEVVQSL 585
++ +L L I +GR++ IL+LV+IC +W + + L + ++Q++
Sbjct: 1917 LVLMLLLRSIMKGREENILLLVLIC-SWLITLLIFLILFTVLIQTV 2051
>TC77916 similar to GP|17979073|gb|AAL49804.1 putative gamma glutamyl
hydrolase {Arabidopsis thaliana}, partial (82%)
Length = 1695
Score = 29.6 bits (65), Expect = 6.5
Identities = 10/24 (41%), Positives = 19/24 (78%)
Frame = -3
Query: 833 VFKFTSGLQSYPCLPTLLQKLILN 856
+FK+ + ++S+PCLPT+ +I+N
Sbjct: 1276 LFKYINFIESFPCLPTISGSVIVN 1205
>AL387383 similar to GP|6682235|gb| unknown protein {Arabidopsis thaliana},
partial (17%)
Length = 487
Score = 29.3 bits (64), Expect = 8.4
Identities = 12/33 (36%), Positives = 20/33 (60%)
Frame = -2
Query: 689 KYFVNWR*FFLLVSLT*WFTCLSISQMRLDWVV 721
KYF W +F + F C+SI++ R++WV+
Sbjct: 354 KYFSLWNSYFTVTK----FICVSIAKPRIEWVI 268
>BQ139285 weakly similar to PIR|T05099|T05 hypothetical protein F28M20.100 -
Arabidopsis thaliana, partial (17%)
Length = 682
Score = 29.3 bits (64), Expect = 8.4
Identities = 12/28 (42%), Positives = 16/28 (56%)
Frame = +1
Query: 417 LSGWSTKGKLACACCNYNTSSTYLKHSH 444
+S T GK+ +C N + S YL HSH
Sbjct: 157 ISSNETVGKIGLSCINMSLSDFYLSHSH 240
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.348 0.153 0.544
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,096,010
Number of Sequences: 36976
Number of extensions: 648826
Number of successful extensions: 6557
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 2692
Number of HSP's successfully gapped in prelim test: 317
Number of HSP's that attempted gapping in prelim test: 3743
Number of HSP's gapped (non-prelim): 3257
length of query: 1065
length of database: 9,014,727
effective HSP length: 106
effective length of query: 959
effective length of database: 5,095,271
effective search space: 4886364889
effective search space used: 4886364889
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0250.10


