

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0238.21
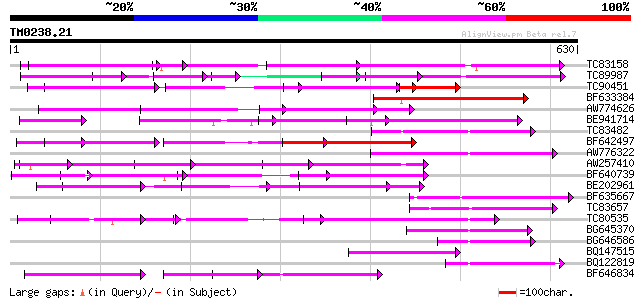
(630 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein... 202 3e-52
TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat ... 134 3e-39
TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13... 104 6e-35
BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30... 139 3e-33
AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.7... 129 4e-30
BE941714 weakly similar to GP|11994734|dbj selenium-binding prot... 124 1e-28
TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-bin... 122 5e-28
BF642497 weakly similar to PIR|H96713|H96 hypothetical protein T... 117 2e-26
AW776322 weakly similar to PIR|H84859|H84 hypothetical protein A... 110 2e-24
AW257410 weakly similar to GP|10177533|db selenium-binding prote... 107 1e-23
BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis... 105 7e-23
BE202961 weakly similar to GP|6016735|gb|A hypothetical protein ... 101 8e-22
BF635667 weakly similar to PIR|C85359|C853 hypothetical protein ... 88 1e-17
TC83657 weakly similar to GP|11994382|dbj|BAB02341. gb|AAB82628.... 86 4e-17
TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9... 84 1e-16
BG645370 weakly similar to PIR|T00405|T004 hypothetical protein ... 83 3e-16
BG646586 weakly similar to GP|10177683|dbj selenium-binding prot... 80 3e-15
BQ147515 weakly similar to PIR|T05355|T05 hypothetical protein F... 79 7e-15
BQ122819 similar to PIR|C84748|C84 hypothetical protein At2g3368... 75 6e-14
BF646834 weakly similar to PIR|A84784|A847 hypothetical protein ... 74 2e-13
>TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein [imported]
- Arabidopsis thaliana, partial (42%)
Length = 1194
Score = 202 bits (514), Expect = 3e-52
Identities = 118/335 (35%), Positives = 180/335 (53%), Gaps = 4/335 (1%)
Frame = +2
Query: 286 GDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVL 345
GD A F + P KN+VSWT +I G+ + E AL F +M + D + A++
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 346 HACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLV 405
ACA+L L G VH ++++ + V NSL++MYA+CG +E + F G+ +++LV
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLV 361
Query: 406 SWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSM 465
SWNS++ F ++G A++A+ FR M G++P+ V++T L CSH GLIDEG F +
Sbjct: 362 SWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADI 541
Query: 466 SSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLG----ACHA 521
+ S ++H C+VD+ R G + EA + KK EV+LG AC
Sbjct: 542 KRDHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPMMPN------EVVLGSLLAACRT 703
Query: 522 HGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGSS 581
GD+ V +Y L P + YV+ SN+Y A G+W A VR+EM ++G++K S
Sbjct: 704 QGDVELAEKVMKYQVELYPGGDSNYVLFSNIYAAVGKWDGASKVRREMKERGLQKNLAFS 883
Query: 582 WIEIRNVVTAFVSGNNSSPYMADISNILYFLEIEM 616
IEI + + FVSG+ I + L L E+
Sbjct: 884 SIEIDSGIHKFVSGDKYHEENDYIYSALELLSFEL 988
Score = 101 bits (251), Expect = 1e-21
Identities = 58/180 (32%), Positives = 98/180 (54%), Gaps = 4/180 (2%)
Frame = +2
Query: 22 GRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAAL 81
G + A KLFD++P ++ V+W +I + Y+++L F M+++ PD + A +
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 82 SACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEV 141
SACA G G +H LV+ +R ++ V NSLIDMY +C AR+VFD M+ N V
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLV 361
Query: 142 TWCSLLFAYANSSLFGMALEVFRSMP----ERVEIAWNTMIAGHARRGEVEACLGLFKEM 197
+W S++ +A + L AL FRSM E +++ + + + G ++ L +F ++
Sbjct: 362 SWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADI 541
Score = 81.3 bits (199), Expect = 1e-15
Identities = 60/234 (25%), Positives = 105/234 (44%), Gaps = 2/234 (0%)
Frame = +2
Query: 159 ALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTE 218
AL++F +P + ++W +I G ++ E L F+EM + PD T A+++AC
Sbjct: 17 ALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISACAN 196
Query: 219 SRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAI 278
+ G VH V+K + ++V NS++ YA+ C A ++F+ N VSWN
Sbjct: 197 LGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSWN-- 370
Query: 279 IDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDN 338
S+IVG+ NG + ALS F M + ++ +
Sbjct: 371 -----------------------------SIIVGFAVNGLADKALSFFRSMKKEGLEPNG 463
Query: 339 LVAGAVLHACASLAILAHGKMVHSCIIR--RGLDKYLFVGNSLVNMYAKCGDLE 390
+ + L AC+ ++ G + + I R R + G LV++Y++ G L+
Sbjct: 464 VSYTSALTACSHAGLIDEGLKIFADIKRDHRNSPRIEHYG-CLVDLYSRAGRLK 622
Score = 62.4 bits (150), Expect = 5e-10
Identities = 51/159 (32%), Positives = 82/159 (51%), Gaps = 3/159 (1%)
Frame = +2
Query: 13 SKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKP 72
S I AR G I AR++FD M R+ V+WN++I ++ GL ++LS F SM+ +P
Sbjct: 278 SLIDMYARCGCIELARQVFDGMSQRNLVSWNSIIVGFAVNGLADKALSFFRSMKKEGLEP 457
Query: 73 DSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVAN--SLIDMYGKCLKPHDARK 130
+ SY++AL+AC+ G I A + +R+S + + L+D+Y + + +A
Sbjct: 458 NGVSYTSALTACSHAGLIDEGLKIFA-DIKRDHRNSPRIEHYGCLVDLYSRAGRLKEAWD 634
Query: 131 VFDEM-ADSNEVTWCSLLFAYANSSLFGMALEVFRSMPE 168
V +M NEV SLL A +A +V + E
Sbjct: 635 VIKKMPMMPNEVVLGSLLAACRTQGDVELAEKVMKYQVE 751
>TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat protein
[imported] - Arabidopsis thaliana, partial (62%)
Length = 1559
Score = 134 bits (338), Expect(2) = 3e-39
Identities = 73/224 (32%), Positives = 118/224 (52%), Gaps = 2/224 (0%)
Frame = +2
Query: 396 FCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLI 455
F + EK+ S+ M+ +HGR EA+ +F EM+ G+ PD+V + G+ CSH GL+
Sbjct: 584 FKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHAGLV 763
Query: 456 DEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNS--YE 513
+EG F+SM E + + H CMVD+LGR G + EA L K S + N +
Sbjct: 764 EEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMS----IKPNDVIWR 931
Query: 514 VLLGACHAHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQG 573
LL AC H +L G E L L Y++L+N+Y + +W + +R ++ ++
Sbjct: 932 SLLSACKVHHNLEIGKIAAENLFMLNQNNSGDYLVLANMYAKAQKWDDVAKIRTKLAERN 1111
Query: 574 VKKVPGSSWIEIRNVVTAFVSGNNSSPYMADISNILYFLEIEMR 617
+ + PG S IE + V FVS + S P I +++ +E +++
Sbjct: 1112LVQTPGFSLIEAKRKVYKFVSQDKSIPQWNIIYEMIHQMEWQVK 1243
Score = 76.3 bits (186), Expect = 3e-14
Identities = 39/113 (34%), Positives = 64/113 (56%), Gaps = 1/113 (0%)
Frame = +1
Query: 347 ACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVS 406
AC+ L ++ G VH + + GL+ + V NSL+NMY KCG+++ + F G+ EK + S
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 407 WNSMLFAFGLHGRANEAMCLFREMVASG-VKPDEVTFTGMLMTCSHLGLIDEG 458
W++++ A NE + L +M + G + +E T +L C+HLG D G
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLG 468
Score = 52.0 bits (123), Expect = 7e-07
Identities = 40/164 (24%), Positives = 69/164 (41%)
Frame = +1
Query: 93 GSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYAN 152
G +H V G + V NSLI+MYGKC + +A VF+ M + + +W +++ A+A
Sbjct: 160 GIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVASWSAIIGAHAC 339
Query: 153 SSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSAL 212
+ WN E + L K E + ++ T +
Sbjct: 340 VEM------------------WN------------ECLMLLGKMSSEGRCRVEESTLVNV 429
Query: 213 MNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLEC 256
++ACT G +HG ++++ + VK S++ Y K C
Sbjct: 430 LSACTHLGSPDLGKCIHGILLRNISELNVVVKTSLIDMYVKSGC 561
Score = 46.2 bits (108), Expect(2) = 3e-39
Identities = 39/167 (23%), Positives = 65/167 (38%), Gaps = 1/167 (0%)
Frame = +1
Query: 225 GCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDAHMK 284
G VHG V K G + V+NS+++ Y K
Sbjct: 160 GIQVHGHVFKMGLEGDVIVQNSLINMYGKC------------------------------ 249
Query: 285 LGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNS-IQLDNLVAGA 343
G+ + A F +K++ SW+++I + L + M+ +++
Sbjct: 250 -GEIKNACDVFNGMDEKSVASWSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVN 426
Query: 344 VLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLE 390
VL AC L GK +H ++R + + V SL++MY K G LE
Sbjct: 427 VLSACTHLGSPDLGKCIHGILLRNISELNVVVKTSLIDMYVKSGCLE 567
Score = 45.8 bits (107), Expect(2) = 9e-11
Identities = 33/120 (27%), Positives = 58/120 (47%), Gaps = 3/120 (2%)
Frame = +1
Query: 13 SKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNS-K 71
S I + G I +A +F+ M ++ +W+A+I A++ + ++ + L L G M +
Sbjct: 223 SLINMYGKCGEIKNACDVFNGMDEKSVASWSAIIGAHACVEMWNECLMLLGKMSSEGRCR 402
Query: 72 PDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGK--CLKPHDAR 129
+ + LSAC G IH +++ + ++ V SLIDMY K CL+ R
Sbjct: 403 VEESTLVNVLSACTHLGSPDLGKCIHGILLRNISELNVVVKTSLIDMYVKSGCLEKRFTR 582
Score = 38.9 bits (89), Expect(2) = 9e-11
Identities = 19/60 (31%), Positives = 32/60 (52%)
Frame = +2
Query: 160 LEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTES 219
L VF++M E+ ++ MI+G A G + L +F EM E PD + + +AC+ +
Sbjct: 575 LRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHA 754
Score = 34.7 bits (78), Expect = 0.11
Identities = 29/138 (21%), Positives = 60/138 (43%), Gaps = 3/138 (2%)
Frame = +2
Query: 15 IVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDS 74
++ ++R + ++F M +++ ++ MI+ + G +++L +F M PD
Sbjct: 536 LICMSRVDVLRKGLRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDD 715
Query: 75 FSYSAALSAC--AGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVF 132
Y SAC AG G ++ ++ ++D+ G+ +A ++
Sbjct: 716 VVYVGVFSACSHAGLVEEGL-QCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELI 892
Query: 133 DEMA-DSNEVTWCSLLFA 149
M+ N+V W SLL A
Sbjct: 893 KSMSIKPNDVIWRSLLSA 946
>TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13600
[imported] - Arabidopsis thaliana, partial (12%)
Length = 1362
Score = 104 bits (259), Expect(2) = 6e-35
Identities = 64/264 (24%), Positives = 120/264 (45%), Gaps = 2/264 (0%)
Frame = +2
Query: 174 WNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVHGFVI 233
WN +I + R + L ++ M + PD++T ++ A ++S + G VH + I
Sbjct: 389 WNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGI 568
Query: 234 KSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDAHMKLGDTQKAFL 293
K G S N+ + I+ + K GD A
Sbjct: 569 KLGLQS-------------------------------NEYCESGFINLYCKAGDFDSAHK 655
Query: 294 AFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASLAI 353
F + + + SW ++I G ++ G A+ +F+DM R+ + D + +V+ AC S+
Sbjct: 656 VFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGD 835
Query: 354 LAHGKMVHSCIIRRGLDKY--LFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSML 411
L +H + + +++ + + NSL++MY KCG ++ + F + ++++ SW SM+
Sbjct: 836 LYLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMI 1015
Query: 412 FAFGLHGRANEAMCLFREMVASGV 435
+ +HG A EA+ F M GV
Sbjct: 1016VGYAMHGHAKEALGCFHCMKGVGV 1087
Score = 78.6 bits (192), Expect = 7e-15
Identities = 67/288 (23%), Positives = 116/288 (40%)
Frame = +2
Query: 39 SVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSACAGGSHHGFGSVIHA 98
S WN +I +Y+ L Q +L ++ SM + PD ++ L A + G +H+
Sbjct: 380 SFNWNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHS 559
Query: 99 LVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYANSSLFGM 158
+ G +S+ + I++Y K A KVFD +++E S
Sbjct: 560 YGIKLGLQSNEYCESGFINLYCKAGDFDSAHKVFD---ENHEPKLGS------------- 691
Query: 159 ALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTE 218
WN +I+G ++ G + +F +M ++PD T ++M+AC
Sbjct: 692 ---------------WNALISGLSQGGLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGS 826
Query: 219 SRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAI 278
D+ +H +V ++ + + S N++
Sbjct: 827 IGDLYLALQLHKYVFQAKTNEWTVILMS-----------------------------NSL 919
Query: 279 IDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMF 326
ID + K G A+ F D+N+ SWTSMIVGY +G+ + AL F
Sbjct: 920 IDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCF 1063
Score = 77.4 bits (189), Expect = 2e-14
Identities = 42/149 (28%), Positives = 73/149 (48%)
Frame = +2
Query: 305 SWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSCI 364
+W ++I YTR + + AL +++ M R + D VL A + + G+ VHS
Sbjct: 386 NWNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYG 565
Query: 365 IRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAM 424
I+ GL + + +N+Y K GD + + F E L SWN+++ G A +A+
Sbjct: 566 IKLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAI 745
Query: 425 CLFREMVASGVKPDEVTFTGMLMTCSHLG 453
+F +M G +PD +T ++ C +G
Sbjct: 746 VVFVDMKRHGFEPDGITMVSVMSACGSIG 832
Score = 70.9 bits (172), Expect = 1e-12
Identities = 46/149 (30%), Positives = 72/149 (47%), Gaps = 2/149 (1%)
Frame = +2
Query: 20 RSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSA 79
++G A K+FDE + +WNA+I+ S GL ++ +F M+ +PD + +
Sbjct: 626 KAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPDGITMVS 805
Query: 80 ALSACAGGSHHGFGSVIHALVVVSGYR--SSLPVANSLIDMYGKCLKPHDARKVFDEMAD 137
+SAC +H V + + + ++NSLIDMYGKC + A +VF M D
Sbjct: 806 VMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEVFATMED 985
Query: 138 SNEVTWCSLLFAYANSSLFGMALEVFRSM 166
N +W S++ YA AL F M
Sbjct: 986 RNVSSWTSMIVGYAMHGHAKEALGCFHCM 1072
Score = 62.0 bits (149), Expect(2) = 6e-35
Identities = 27/67 (40%), Positives = 42/67 (62%)
Frame = +1
Query: 434 GVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAE 493
GVKP+ VTF G+L C H G + EG +F M + +G++ + H CMVD+LGR G +
Sbjct: 1084 GVKPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQHYGCMVDLLGRAGLFDD 1263
Query: 494 AQSLAKK 500
A+ + ++
Sbjct: 1264 ARRMVEE 1284
>BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30 -
Arabidopsis thaliana, partial (22%)
Length = 595
Score = 139 bits (351), Expect = 3e-33
Identities = 68/177 (38%), Positives = 110/177 (61%), Gaps = 5/177 (2%)
Frame = +1
Query: 405 VSWNSMLFAFGLHGRANEAMCLFREMVASG-----VKPDEVTFTGMLMTCSHLGLIDEGF 459
++WN ++ A+G+HG+ EA+ LFR MV G ++P+EVT+ + + SH G++DEG
Sbjct: 1 ITWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGL 180
Query: 460 AFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGAC 519
F +M ++ G+ DH AC+VD+LGR G + EA +L K ++ + +++ LLGAC
Sbjct: 181 NLFYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKT-MPSNMKKVDAWSSLLGAC 357
Query: 520 HAHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKK 576
H +L G + L L+P YV+LSN+Y ++G W +A VRK+M ++GV+K
Sbjct: 358 KIHQNLEIGEIAAKNLFVLDPNVASYYVLLSNIYSSAGLWDQAIDVRKKMKEKGVRK 528
>AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.70 -
Arabidopsis thaliana, partial (12%)
Length = 703
Score = 129 bits (323), Expect = 4e-30
Identities = 77/266 (28%), Positives = 134/266 (49%), Gaps = 2/266 (0%)
Frame = +3
Query: 146 LLFAYANSSLFGMALEVFRSMP--ERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQ 203
L+ YA A +F+ + + + W M+ G+A+ G+ + F+ M +
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 204 PDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEM 263
+Q+TF ++ AC+ +G VHGF++KSG+ S + V+++++ YAK
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKC--------- 335
Query: 264 FNSFGAFNQVSWNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELAL 323
GD + A + D ++VSW S++VG+ R+G E AL
Sbjct: 336 ----------------------GDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGLEEEAL 449
Query: 324 SMFLDMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMY 383
+F +M ++++D+ +VL+ C +I + K VH II+ G + Y V N+LV+MY
Sbjct: 450 RLFKNMHGRNMKIDDYTFPSVLNCCVVGSI--NPKSVHGLIIKTGFENYKLVSNALVDMY 623
Query: 384 AKCGDLEGSALAFCGILEKDLVSWNS 409
AK GD++ + F +LEKD++SW S
Sbjct: 624 AKTGDMDCAYTVFEKMLEKDVISWTS 701
Score = 111 bits (277), Expect = 1e-24
Identities = 58/174 (33%), Positives = 95/174 (54%), Gaps = 2/174 (1%)
Frame = +3
Query: 278 IIDAHMKLGDTQKAFLAFQ--QAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQ 335
++D + K +A F+ + KN V WT+M+ GY +NG+G A+ F M ++
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 336 LDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALA 395
+ +L AC+S+ G+ VH I++ G ++V ++LV+MYAKCGDL+ +
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNM 362
Query: 396 FCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTC 449
+ + D+VSWNS++ F HG EA+ LF+ M +K D+ TF +L C
Sbjct: 363 LETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCC 524
Score = 100 bits (249), Expect = 2e-21
Identities = 68/276 (24%), Positives = 117/276 (41%)
Frame = +3
Query: 33 EMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSACAGGSHHGF 92
E ++ V W AM+T Y+ G +++ F M + + +++ L+AC+ F
Sbjct: 66 EFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCF 245
Query: 93 GSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYAN 152
G +H +V SG+ S++ V ++L+DMY KC +A+ + + M D + V+W SL+
Sbjct: 246 GEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNMLETMEDDDVVSWNSLM----- 410
Query: 153 SSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSAL 212
G R G E L LFK M + D +TF ++
Sbjct: 411 --------------------------VGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSV 512
Query: 213 MNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQ 272
+N C + VHG +IK+G+ + V N+++ YA
Sbjct: 513 LNCCVVG--SINPKSVHGLIIKTGFENYKLVSNALVDMYA-------------------- 626
Query: 273 VSWNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTS 308
K GD A+ F++ +K+++SWTS
Sbjct: 627 -----------KTGDMDCAYTVFEKMLEKDVISWTS 701
>BE941714 weakly similar to GP|11994734|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (10%)
Length = 619
Score = 124 bits (310), Expect = 1e-28
Identities = 68/198 (34%), Positives = 104/198 (52%), Gaps = 3/198 (1%)
Frame = +2
Query: 375 VGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASG 434
+ SL++MYAKCG+LE + F + +D+V WN+M+ +HG A+ LF +M G
Sbjct: 20 LSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVG 199
Query: 435 VKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEA 494
VKPD++TF + CS+ G+ EG M S + + +H C+VD+L R G EA
Sbjct: 200 VKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSRAGLFEEA 379
Query: 495 QSLAKKYSKT--SGARTNSYEVLLGACHAHGDLGTGSSVGEYLKTLEPEKEVG-YVMLSN 551
+ +K + + T ++ L AC HG+ E + L+ G YV+LSN
Sbjct: 380 MVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVLQLDNHIHSGVYVLLSN 559
Query: 552 LYCASGQWKEAEIVRKEM 569
LY ASG+ +A V+ M
Sbjct: 560 LYAASGKHNDARRVKDMM 613
Score = 59.7 bits (143), Expect = 3e-09
Identities = 49/165 (29%), Positives = 73/165 (43%), Gaps = 12/165 (7%)
Frame = +2
Query: 145 SLLFAYANSSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQP 204
SLL YA +A +F SM R + WN MI+G A G+ + L LF +M + +P
Sbjct: 29 SLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVGVKP 208
Query: 205 DQWTFSALMNACTESRDMLYG-------CMVHGFVIKSGWSSAMEVKNSILSFYAKLECP 257
D TF A+ AC+ S G C V+ V KS E ++ ++
Sbjct: 209 DDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKS------EHYGCLVDLLSRAGLF 370
Query: 258 SDAMEMFNSF-----GAFNQVSWNAIIDAHMKLGDTQKAFLAFQQ 297
+AM M G+ ++W A + A G+TQ A LA ++
Sbjct: 371 EEAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEK 505
Score = 52.0 bits (123), Expect = 7e-07
Identities = 38/154 (24%), Positives = 73/154 (46%), Gaps = 7/154 (4%)
Frame = +2
Query: 277 AIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQL 336
+++D + K G+ + A F +++V W +MI G +G+G+ AL +F DM + ++
Sbjct: 29 SLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVGVKP 208
Query: 337 DNLVAGAVLHACASLAILAHGKMV--HSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSAL 394
D++ AV AC+ + G M+ C + + K G LV++ ++ G E + +
Sbjct: 209 DDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYG-CLVDLLSRAGLFEEAMV 385
Query: 395 AFCGIL-----EKDLVSWNSMLFAFGLHGRANEA 423
I ++ ++W + L A HG A
Sbjct: 386 MIRKITNSWNGSEETLAWRAFLSACCNHGETQLA 487
Score = 51.2 bits (121), Expect = 1e-06
Identities = 28/75 (37%), Positives = 42/75 (55%)
Frame = +2
Query: 11 TTSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNS 70
+TS + A+ G + A++LFD M RD V WNAMI+ + G + +L LF M
Sbjct: 23 STSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVGV 202
Query: 71 KPDSFSYSAALSACA 85
KPD ++ A +AC+
Sbjct: 203KPDDITFIAVFTACS 247
>TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-binding
protein-like {Arabidopsis thaliana}, partial (10%)
Length = 895
Score = 122 bits (305), Expect = 5e-28
Identities = 66/182 (36%), Positives = 94/182 (51%)
Frame = +2
Query: 403 DLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFF 462
DLVSWN+++ + HG A A+ F M G PDEVTF +L C H GL++EG F
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVGT-PDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 463 RSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGACHAH 522
M +++G+ M+H +CMVD+ GR G EA++L K + LL AC H
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVL--WGALLAACGLH 352
Query: 523 GDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGSSW 582
+L G E ++ LE V Y +LS + G W +R M ++G+KK SW
Sbjct: 353 SNLELGEYAAERIRRLESSHPVSYSVLSKIQGEKGVWSSVNELRDTMKERGIKKQTAISW 532
Query: 583 IE 584
+E
Sbjct: 533 VE 538
Score = 41.6 bits (96), Expect = 0.001
Identities = 33/118 (27%), Positives = 54/118 (44%), Gaps = 2/118 (1%)
Frame = +2
Query: 302 NIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASLAILAHG-KMV 360
++VSW ++I GY +G AL F D + D + VL AC ++ G K
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEF-DRMKVVGTPDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 361 HSCIIRRGLDKYLFVGNSLVNMYAKCGDL-EGSALAFCGILEKDLVSWNSMLFAFGLH 417
+ + G+ + + +V++Y + G E L E D+V W ++L A GLH
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLH 352
>BF642497 weakly similar to PIR|H96713|H96 hypothetical protein T6L1.11
[imported] - Arabidopsis thaliana, partial (20%)
Length = 461
Score = 117 bits (292), Expect = 2e-26
Identities = 55/149 (36%), Positives = 92/149 (60%)
Frame = +3
Query: 304 VSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSC 363
+SWTSMI G+T+NG A+ +F +M ++Q+D G+VL AC + L GK VH+
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAY 185
Query: 364 IIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEA 423
IIR +FV ++LV+MY KC +++ + F + K++VSW +ML +G +G + EA
Sbjct: 186 IIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEA 365
Query: 424 MCLFREMVASGVKPDEVTFTGMLMTCSHL 452
+ F +M G++PD+ T ++ +C++L
Sbjct: 366 VKTFSDMQKYGIEPDDFTLGSVISSCANL 452
Score = 89.7 bits (221), Expect = 3e-18
Identities = 54/183 (29%), Positives = 90/183 (48%)
Frame = +3
Query: 172 IAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVHGF 231
I+W +MI G + G + +F+EM Q DQ+TF +++ AC + G VH +
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAY 185
Query: 232 VIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDAHMKLGDTQKA 291
+I++ + N F A +A++D + K + + A
Sbjct: 186 IIRTDYKD-------------------------NIFVA------SALVDMYCKCKNIKSA 272
Query: 292 FLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASL 351
F++ KN+VSWT+M+VGY +NG E A+ F DM + I+ D+ G+V+ +CA+L
Sbjct: 273 EAVFKKMTCKNVVSWTAMLVGYGQNGYSEEAVKTFSDMQKYGIEPDDFTLGSVISSCANL 452
Query: 352 AIL 354
A L
Sbjct: 453 ASL 461
Score = 87.0 bits (214), Expect = 2e-17
Identities = 39/128 (30%), Positives = 73/128 (56%)
Frame = +3
Query: 39 SVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSACAGGSHHGFGSVIHA 98
S++W +MIT ++ GL + ++ +F M++ N + D +++ + L+AC G G +HA
Sbjct: 3 SISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHA 182
Query: 99 LVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYANSSLFGM 158
++ + Y+ ++ VA++L+DMY KC A VF +M N V+W ++L Y +
Sbjct: 183 YIIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEE 362
Query: 159 ALEVFRSM 166
A++ F M
Sbjct: 363 AVKTFSDM 386
Score = 43.1 bits (100), Expect = 3e-04
Identities = 19/78 (24%), Positives = 40/78 (50%)
Frame = +3
Query: 8 LFQTTSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRI 67
+F ++ + + I A +F +M ++ V+W AM+ Y G ++++ F M+
Sbjct: 213 IFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEAVKTFSDMQK 392
Query: 68 SNSKPDSFSYSAALSACA 85
+PD F+ + +S+CA
Sbjct: 393 YGIEPDDFTLGSVISSCA 446
Score = 40.4 bits (93), Expect = 0.002
Identities = 18/54 (33%), Positives = 30/54 (55%)
Frame = +3
Query: 405 VSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEG 458
+SW SM+ F +G +A+ +FREM ++ D+ TF +L C + + EG
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEG 167
>AW776322 weakly similar to PIR|H84859|H84 hypothetical protein At2g42920
[imported] - Arabidopsis thaliana, partial (27%)
Length = 633
Score = 110 bits (275), Expect = 2e-24
Identities = 63/208 (30%), Positives = 108/208 (51%), Gaps = 1/208 (0%)
Frame = +1
Query: 402 KDLVSWNSMLFAFGLHGRANEAMCLFREMVASGV-KPDEVTFTGMLMTCSHLGLIDEGFA 460
+ L WNS++ ++G EA F ++ +S + KPD V+F G+L C HLG I++
Sbjct: 16 RGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKARD 195
Query: 461 FFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGACH 520
+F M +++ + + H C+VD+LG+ G + EA+ L K A + LL +C
Sbjct: 196 YFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAII--WGSLLSSCR 369
Query: 521 AHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGS 580
H ++ + + L P GYV++SN++ AS +++EA R M + +K PG
Sbjct: 370 KHRNVQIARRAAQRVYELNPSDASGYVLMSNVHAASNKFEEAIEQRLLMKENLTEKEPGC 549
Query: 581 SWIEIRNVVTAFVSGNNSSPYMADISNI 608
S IE+ V F++G P +I ++
Sbjct: 550 SSIELYGEVHEFIAGGRLHPKTQEIYHL 633
Score = 33.5 bits (75), Expect = 0.25
Identities = 29/129 (22%), Positives = 56/129 (42%), Gaps = 7/129 (5%)
Frame = +1
Query: 296 QQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNS-IQLDNLVAGAVLHACASLAIL 354
+ P + + W S+I+G NG+ A F + + ++ D++ VL AC L +
Sbjct: 1 ETCPRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAI 180
Query: 355 AHGK-----MVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGI-LEKDLVSWN 408
+ M++ I + Y +V++ + G LE + G+ L+ D + W
Sbjct: 181 NKARDYFELMMNKYEIEPSIKHY----TCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWG 348
Query: 409 SMLFAFGLH 417
S+L + H
Sbjct: 349 SLLSSCRKH 375
Score = 32.3 bits (72), Expect = 0.56
Identities = 36/168 (21%), Positives = 68/168 (40%), Gaps = 3/168 (1%)
Frame = +1
Query: 35 PDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNS-KPDSFSYSAALSACAG-GSHHGF 92
P R WN++I + G +++ F + S KPDS S+ L+AC G+ +
Sbjct: 10 PRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKA 189
Query: 93 GSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEM-ADSNEVTWCSLLFAYA 151
++ S+ ++D+ G+ +A ++ M + + W SLL
Sbjct: 190 RDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLL---- 357
Query: 152 NSSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCE 199
+S +++ R +RV + +G+ V A F+E E
Sbjct: 358 SSCRKHRNVQIARRAAQRVYELNPSDASGYVLMSNVHAASNKFEEAIE 501
>AW257410 weakly similar to GP|10177533|db selenium-binding protein-like
{Arabidopsis thaliana}, partial (21%)
Length = 724
Score = 107 bits (268), Expect = 1e-23
Identities = 64/201 (31%), Positives = 103/201 (50%), Gaps = 2/201 (0%)
Frame = +1
Query: 139 NEVTWCSLLFAYANSSLFGM--ALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKE 196
+++T LL YA+ + A VF + + WNTMI ++ + E L L+ +
Sbjct: 70 HKLTVSRLLTTYASMEFSNLTYARMVFDRISSPNTVMWNTMIRAYSNSNDPEEALLLYHQ 249
Query: 197 MCESLYQPDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLEC 256
M + +TF L+ AC+ + +H +IK G+ S + NS+L YA
Sbjct: 250 MLHHSIPHNAYTFPFLLKACSALSALAETHQIHVQIIKRGFGSEVYATNSLLRVYAISGS 429
Query: 257 PSDAMEMFNSFGAFNQVSWNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRN 316
A +F+ + + VSWN +ID ++K G+ + A+ FQ P+KN++SWTSMIVG+ R
Sbjct: 430 IKSAHVLFDLLPSRDIVSWNTMIDGYIKCGNVEMAYKIFQAMPEKNVISWTSMIVGFVRT 609
Query: 317 GNGELALSMFLDMTRNSIQLD 337
G + ALS+ M I+ D
Sbjct: 610 GMHKKALSLLQQMLVARIKPD 672
Score = 92.8 bits (229), Expect = 4e-19
Identities = 54/199 (27%), Positives = 102/199 (51%), Gaps = 3/199 (1%)
Frame = +1
Query: 11 TTSKIVSLARS---GRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRI 67
T S++++ S + +AR +FD + ++V WN MI AYS+ +++L L+ M
Sbjct: 79 TVSRLLTTYASMEFSNLTYARMVFDRISSPNTVMWNTMIRAYSNSNDPEEALLLYHQMLH 258
Query: 68 SNSKPDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHD 127
+ +++++ L AC+ S IH ++ G+ S + NSL+ +Y
Sbjct: 259 HSIPHNAYTFPFLLKACSALSALAETHQIHVQIIKRGFGSEVYATNSLLRVYAISGSIKS 438
Query: 128 ARKVFDEMADSNEVTWCSLLFAYANSSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEV 187
A +FD + + V+W +++ Y MA ++F++MPE+ I+W +MI G R G
Sbjct: 439 AHVLFDLLPSRDIVSWNTMIDGYIKCGNVEMAYKIFQAMPEKNVISWTSMIVGFVRTGMH 618
Query: 188 EACLGLFKEMCESLYQPDQ 206
+ L L ++M + +PD+
Sbjct: 619 KKALSLLQQMLVARIKPDK 675
Score = 82.0 bits (201), Expect = 6e-16
Identities = 49/185 (26%), Positives = 92/185 (49%)
Frame = +1
Query: 281 AHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLV 340
A M+ + A + F + N V W +MI Y+ + + E AL ++ M +SI +
Sbjct: 106 ASMEFSNLTYARMVFDRISSPNTVMWNTMIRAYSNSNDPEEALLLYHQMLHHSIPHNAYT 285
Query: 341 AGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGIL 400
+L AC++L+ LA +H II+RG ++ NSL+ +YA G ++ + + F +
Sbjct: 286 FPFLLKACSALSALAETHQIHVQIIKRGFGSEVYATNSLLRVYAISGSIKSAHVLFDLLP 465
Query: 401 EKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFA 460
+D+VSWN+M+ + G A +F+ M V +++T M++ G+ + +
Sbjct: 466 SRDIVSWNTMIDGYIKCGNVEMAYKIFQAMPEKNV----ISWTSMIVGFVRTGMHKKALS 633
Query: 461 FFRSM 465
+ M
Sbjct: 634 LLQQM 648
Score = 43.5 bits (101), Expect = 2e-04
Identities = 24/64 (37%), Positives = 33/64 (51%)
Frame = +1
Query: 6 SYLFQTTSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSM 65
S ++ T S + A SG I A LFD +P RD V+WN MI Y G + + +F +M
Sbjct: 376 SEVYATNSLLRVYAISGSIKSAHVLFDLLPSRDIVSWNTMIDGYIKCGNVEMAYKIFQAM 555
Query: 66 RISN 69
N
Sbjct: 556 PEKN 567
>BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis thaliana
DNA chromosome 4 BAC clone F4I10; vegetative storage
protein., partial (14%)
Length = 444
Score = 105 bits (261), Expect = 7e-23
Identities = 50/145 (34%), Positives = 86/145 (58%), Gaps = 1/145 (0%)
Frame = +1
Query: 322 ALSMFLDMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVN 381
AL++F M I+ D+ +V+ A A L++ K +H IR +D +FV +LV+
Sbjct: 10 ALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALVD 189
Query: 382 MYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMV-ASGVKPDEV 440
MYAKCG +E + F + E+ +++WN+M+ +G HG A+ LF +M + +KP+++
Sbjct: 190 MYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPNDI 369
Query: 441 TFTGMLMTCSHLGLIDEGFAFFRSM 465
TF ++ CSH G ++EG +F+ M
Sbjct: 370 TFLSVISACSHSGFVEEGLYYFKIM 444
Score = 72.4 bits (176), Expect = 5e-13
Identities = 45/146 (30%), Positives = 78/146 (52%), Gaps = 5/146 (3%)
Frame = +1
Query: 57 QSLSLFGSMRISNSKPDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLI 116
++L+LF +M+ KPDSF++ + ++A A S IH L + + +++ VA +L+
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 117 DMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYANSSLFGMALEVFRSMPERV-----E 171
DMY KC AR++FD M + + +TW +++ Y L AL++F M +
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 172 IAWNTMIAGHARRGEVEACLGLFKEM 197
I + ++I+ + G VE L FK M
Sbjct: 367 ITFLSVISACSHSGFVEEGLYYFKIM 444
Score = 59.3 bits (142), Expect = 4e-09
Identities = 35/91 (38%), Positives = 52/91 (56%), Gaps = 1/91 (1%)
Frame = +1
Query: 3 SMRSYLFQTTSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLF 62
+M + +F T+ + A+ G I AR+LFD M +R + WNAMI Y GL + +L LF
Sbjct: 148 NMDTNVFVATALVDMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLF 327
Query: 63 GSMRISNS-KPDSFSYSAALSACAGGSHHGF 92
M+ S KP+ ++ + +SAC SH GF
Sbjct: 328 DDMQNEASLKPNDITFLSVISAC---SHSGF 411
Score = 51.6 bits (122), Expect = 9e-07
Identities = 39/172 (22%), Positives = 71/172 (40%), Gaps = 1/172 (0%)
Frame = +1
Query: 187 VEACLGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNS 246
V L LF M +PD +TF +++ A + +HG I++ + + V +
Sbjct: 1 VNEALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATA 180
Query: 247 ILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSW 306
++ YAK A E+F+ + ++WNA+ID
Sbjct: 181 LVDMYAKCGAIETARELFDMMQERHVITWNAMID-------------------------- 282
Query: 307 TSMIVGYTRNGNGELALSMFLDMTRN-SIQLDNLVAGAVLHACASLAILAHG 357
GY +G G+ AL +F DM S++ +++ +V+ AC+ + G
Sbjct: 283 -----GYGTHGLGKAALDLFDDMQNEASLKPNDITFLSVISACSHSGFVEEG 423
Score = 34.3 bits (77), Expect = 0.15
Identities = 19/77 (24%), Positives = 36/77 (46%)
Frame = +1
Query: 421 NEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVAC 480
NEA+ LF M + G+KPD TF ++ + L + + + ++ + +
Sbjct: 4 NEALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQA-KWIHGLAIRTNMDTNVFVATA 180
Query: 481 MVDMLGRGGYVAEAQSL 497
+VDM + G + A+ L
Sbjct: 181 LVDMYAKCGAIETAREL 231
>BE202961 weakly similar to GP|6016735|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (14%)
Length = 613
Score = 101 bits (252), Expect = 8e-22
Identities = 58/234 (24%), Positives = 106/234 (44%)
Frame = +2
Query: 191 LGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSF 250
L L EM + +P+ T +++ AC ++ G +H + G+ V
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVST----- 166
Query: 251 YAKLECPSDAMEMFNSFGAFNQVSWNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMI 310
A++D +MK +KA F + P K++++W +
Sbjct: 167 --------------------------ALMDMYMKCFSPEKAVDIFNRMPKKDVIAWAVLF 268
Query: 311 VGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLD 370
GY NG ++ +F +M + + D + +L + L IL H+ +I+ G +
Sbjct: 269 SGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELGILQQAVCFHAFVIKNGFE 448
Query: 371 KYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAM 424
F+G SL+ +YAKC +E + F G+ KD+V+W+S++ A+G HG+ EA+
Sbjct: 449 NNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIAAYGFHGQGEEAL 610
Score = 87.0 bits (214), Expect = 2e-17
Identities = 61/233 (26%), Positives = 100/233 (42%)
Frame = +2
Query: 59 LSLFGSMRISNSKPDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDM 118
L L M KP+ + + L ACA S+ G IH L V G+ V+ +L+DM
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 119 YGKCLKPHDARKVFDEMADSNEVTWCSLLFAYANSSLFGMALEVFRSMPERVEIAWNTMI 178
Y KC P A +++F MP++ IAW +
Sbjct: 182 YMKCFSPEKA-------------------------------VDIFNRMPKKDVIAWAVLF 268
Query: 179 AGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWS 238
+G+A G V + +F+ M S +PD ++ +E + H FVIK+G+
Sbjct: 269 SGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELGILQQAVCFHAFVIKNGFE 448
Query: 239 SAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDAHMKLGDTQKA 291
+ + S++ YAK DA ++F + V+W++II A+ G ++A
Sbjct: 449 NNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIAAYGFHGQGEEA 607
Score = 77.8 bits (190), Expect = 1e-14
Identities = 39/139 (28%), Positives = 73/139 (52%)
Frame = +2
Query: 323 LSMFLDMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNM 382
L + +M I+ + + +VL ACA ++ L G +H + G + V +L++M
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 383 YAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTF 442
Y KC E + F + +KD+++W + + +G +E+M +FR M++SG +PD +
Sbjct: 182 YMKCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIAL 361
Query: 443 TGMLMTCSHLGLIDEGFAF 461
+L T S LG++ + F
Sbjct: 362 VKILTTVSELGILQQAVCF 418
Score = 75.5 bits (184), Expect = 6e-14
Identities = 37/121 (30%), Positives = 66/121 (53%)
Frame = +2
Query: 30 LFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSACAGGSH 89
+F+ MP +D +AW + + Y+ G+ +S+ +F +M S ++PD+ + L+ +
Sbjct: 218 IFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELGI 397
Query: 90 HGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFA 149
HA V+ +G+ ++ + SLI++Y KC DA KVF M + VTW S++ A
Sbjct: 398 LQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIAA 577
Query: 150 Y 150
Y
Sbjct: 578 Y 580
>BF635667 weakly similar to PIR|C85359|C853 hypothetical protein AT4g30700
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 87.8 bits (216), Expect = 1e-17
Identities = 56/182 (30%), Positives = 83/182 (44%)
Frame = +3
Query: 445 MLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKT 504
++ TC GL+D+G F SM S++GL +H ACMVD+ R G + EA L ++ K+
Sbjct: 15 VVATC---GLLDQGKKIFGSMISDYGLEPTAEHYACMVDLFSRCGRLEEALQLLERM-KS 182
Query: 505 SGARTNSYEVLLGACHAHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEI 564
S + + LL C H ++ G +L LEP YV L +Y + G
Sbjct: 183 SSVTGSMWGALLAGCVMHQNVKIGEVAAHHLFQLEPNNTSNYVALWGIYQSRGMVLGVST 362
Query: 565 VRKEMLDQGVKKVPGSSWIEIRNVVTAFVSGNNSSPYMADISNILYFLEIEMRHTRPINF 624
+ +M D G+ K PG WI I F G+ S P I +Y + + T +
Sbjct: 363 IXGKMRDLGLVKTPGCXWINIAGRAHKFYQGDLSHPLSHIICKRVYEIXXTLLSTNDLGA 542
Query: 625 DI 626
I
Sbjct: 543 HI 548
>TC83657 weakly similar to GP|11994382|dbj|BAB02341.
gb|AAB82628.1~gene_id:MIL23.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1299
Score = 85.9 bits (211), Expect = 4e-17
Identities = 53/165 (32%), Positives = 80/165 (48%), Gaps = 1/165 (0%)
Frame = +3
Query: 445 MLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKT 504
+L C+H GL+ E M E+G+ G+ H CMVD+LGR G + EA L K+
Sbjct: 84 VLSACAHGGLMSEALEVISKME-EYGIEMGIRHYGCMVDLLGRAGKLKEAYELIKRMPMK 260
Query: 505 SGARTNSYEVLLGACHAHGDLGTGSSVGEYLKTLEPEKEVGY-VMLSNLYCASGQWKEAE 563
++GAC H D+ V + + + V+LSN+Y AS +W++AE
Sbjct: 261 PNETVLG--AMIGACWIHSDMKMAEQVMKMIGADSAACVNSHNVLLSNIYAASEKWEKAE 434
Query: 564 IVRKEMLDQGVKKVPGSSWIEIRNVVTAFVSGNNSSPYMADISNI 608
++R M+D G +K+PG S I + N S M +S I
Sbjct: 435 MIRSSMVDGGSEKIPGYSSIILSNSAVDLSISKEISRQMNGLSAI 569
>TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9O13.24
{Arabidopsis thaliana}, partial (55%)
Length = 1727
Score = 84.3 bits (207), Expect = 1e-16
Identities = 47/218 (21%), Positives = 95/218 (43%)
Frame = +2
Query: 327 LDMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKC 386
L++ I+ D + C + K VH ++ + N ++ MY C
Sbjct: 248 LELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNC 427
Query: 387 GDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGML 446
+ + F + +++ SW+ M+ + +E + LF +M G++ T +L
Sbjct: 428 KSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVL 607
Query: 447 MTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSG 506
C +++ + + SM S++G+ G++H ++D+LG+ GY+ EA+ ++
Sbjct: 608 SACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLPFEPT 787
Query: 507 ARTNSYEVLLGACHAHGDLGTGSSVGEYLKTLEPEKEV 544
+E L HGD+ V E + +L+P K V
Sbjct: 788 VTV--FETLKNYARIHGDVDLEDHVEELIVSLDPSKAV 895
Score = 65.1 bits (157), Expect = 8e-11
Identities = 42/145 (28%), Positives = 67/145 (45%), Gaps = 1/145 (0%)
Frame = +2
Query: 46 ITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSACAGGSHHGFGSVIHALVVVSGY 105
+T + G +++L L K D+ + C +H + S +
Sbjct: 209 LTRFCQEGKVKEALELMEK----GIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTF 376
Query: 106 RSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYANSSLFGMALEVFRS 165
RS + N +I+MYG C DAR+VFD M + N +W ++ YANS++ L++F
Sbjct: 377 RSDFKMHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQ 556
Query: 166 MPE-RVEIAWNTMIAGHARRGEVEA 189
M E +EI TM+A + G EA
Sbjct: 557 MNELGLEITSETMLAVLSACGSAEA 631
Score = 50.4 bits (119), Expect = 2e-06
Identities = 39/168 (23%), Positives = 70/168 (41%)
Frame = +2
Query: 183 RRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAME 242
+ G+V+ L E+ E + D F L + C +S+ + VH + ++S + S +
Sbjct: 224 QEGKVKEAL----ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFK 391
Query: 243 VKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDAHMKLGDTQKAFLAFQQAPDKN 302
+ N ++ Y + +DA +F+ HM P++N
Sbjct: 392 MHNKVIEMYGNCKSMTDARRVFD----------------HM---------------PNRN 478
Query: 303 IVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACAS 350
+ SW MI GY + G+ L +F M +++ + AVL AC S
Sbjct: 479 MDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLSACGS 622
Score = 42.7 bits (99), Expect = 4e-04
Identities = 35/150 (23%), Positives = 68/150 (45%), Gaps = 7/150 (4%)
Frame = +2
Query: 9 FQTTSKIVSLARSGR-ICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRI 67
F+ +K++ + + + + AR++FD MP+R+ +W+ MI Y++ + + L LF M
Sbjct: 386 FKMHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNE 565
Query: 68 SNSKPDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVA------NSLIDMYGK 121
+ S + A LSAC +V A + + +S + L+D+ G+
Sbjct: 566 LGLEITSETMLAVLSACGSAE-----AVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQ 730
Query: 122 CLKPHDARKVFDEMADSNEVTWCSLLFAYA 151
+A + +++ VT L YA
Sbjct: 731 SGYLKEAEEFIEQLPFEPTVTVFETLKNYA 820
>BG645370 weakly similar to PIR|T00405|T004 hypothetical protein At2g44880
[imported] - Arabidopsis thaliana, partial (11%)
Length = 784
Score = 83.2 bits (204), Expect = 3e-16
Identities = 45/140 (32%), Positives = 70/140 (49%)
Frame = -2
Query: 442 FTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKY 501
FT +L C+H G ID+G +F M + +GLS +D AC++D+ R G + +A+ L ++
Sbjct: 783 FTAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEM 604
Query: 502 SKTSGARTNSYEVLLGACHAHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKE 561
+ L AC +GD+ G L +EP Y+ L+++Y G W E
Sbjct: 603 PYDPNCII--WSSFLSACKIYGDVELGREAAIQLIKMEPCNAAPYLTLAHIYTTKGLWNE 430
Query: 562 AEIVRKEMLDQGVKKVPGSS 581
A VR M + +K PG S
Sbjct: 429 ASEVRSLMQQRVKRKPPGWS 370
>BG646586 weakly similar to GP|10177683|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (9%)
Length = 687
Score = 79.7 bits (195), Expect = 3e-15
Identities = 41/109 (37%), Positives = 65/109 (59%)
Frame = -3
Query: 476 DHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGACHAHGDLGTGSSVGEYL 535
+H MVD+LGR G + +A SL K+ S ++ A T + LL AC +G++ G +L
Sbjct: 649 EHYTGMVDLLGRAGQLEKAYSLIKENSTSADATT--WGSLLAACRVYGNVELGEIAARHL 476
Query: 536 KTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGSSWIE 584
++P YV+L+N Y ++ +W+ AE V+K M +G+KK G SWI+
Sbjct: 475 FEIDPTDSGNYVLLANTYASNDKWERAEEVKKLMSKKGMKKPSGYSWIQ 329
>BQ147515 weakly similar to PIR|T05355|T05 hypothetical protein F8B4.150 -
Arabidopsis thaliana, partial (23%)
Length = 427
Score = 78.6 bits (192), Expect = 7e-15
Identities = 41/124 (33%), Positives = 67/124 (53%)
Frame = +2
Query: 377 NSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVK 436
N ++ MY +CG ++ + F + E+DL + M+ +G A +++ LF + SG+K
Sbjct: 44 NGILEMYFQCGSVDDAVNVFKNMTERDLTTICIMIKQLAKNGFAEDSIDLFTQFKRSGLK 223
Query: 437 PDEVTFTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQS 496
PD F G+ C LG I EG F SMS ++ + M+H +VDM+G G++ EA
Sbjct: 224 PDGQMFIGVFGACXMLGDIVEGMLHFESMSRDYDIVPTMEHYVSLVDMIGSIGHLDEALE 403
Query: 497 LAKK 500
+K
Sbjct: 404 FIEK 415
Score = 38.9 bits (89), Expect = 0.006
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 2/123 (1%)
Frame = +2
Query: 276 NAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQ 335
N I++ + + G A F+ ++++ + MI +NG E ++ +F R+ ++
Sbjct: 44 NGILEMYFQCGSVDDAVNVFKNMTERDLTTICIMIKQLAKNGFAEDSIDLFTQFKRSGLK 223
Query: 336 LDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGN--SLVNMYAKCGDLEGSA 393
D + V AC L + G M+H + R D + + SLV+M G L+ A
Sbjct: 224 PDGQMFIGVFGACXMLGDIVEG-MLHFESMSRDYDIVPTMEHYVSLVDMIGSIGHLD-EA 397
Query: 394 LAF 396
L F
Sbjct: 398 LEF 406
Score = 33.9 bits (76), Expect = 0.19
Identities = 27/91 (29%), Positives = 42/91 (45%), Gaps = 1/91 (1%)
Frame = +2
Query: 140 EVTWCS-LLFAYANSSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMC 198
+V+ C+ +L Y A+ VF++M ER MI A+ G E + LF +
Sbjct: 29 KVSTCNGILEMYFQCGSVDDAVNVFKNMTERDLTTICIMIKQLAKNGFAEDSIDLFTQFK 208
Query: 199 ESLYQPDQWTFSALMNACTESRDMLYGCMVH 229
S +PD F + AC D++ G M+H
Sbjct: 209 RSGLKPDGQMFIGVFGACXMLGDIVEG-MLH 298
Score = 32.0 bits (71), Expect = 0.74
Identities = 18/63 (28%), Positives = 26/63 (40%)
Frame = +2
Query: 22 GRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAAL 81
G + A +F M +RD MI + G + S+ LF + S KPD +
Sbjct: 74 GSVDDAVNVFKNMTERDLTTICIMIKQLAKNGFAEDSIDLFTQFKRSGLKPDGQMFIGVF 253
Query: 82 SAC 84
AC
Sbjct: 254 GAC 262
>BQ122819 similar to PIR|C84748|C84 hypothetical protein At2g33680 [imported]
- Arabidopsis thaliana, partial (15%)
Length = 771
Score = 75.5 bits (184), Expect = 6e-14
Identities = 47/132 (35%), Positives = 69/132 (51%)
Frame = +2
Query: 485 LGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGACHAHGDLGTGSSVGEYLKTLEPEKEV 544
L R G + EA+ + + G + +LLGAC H + G GE L L +
Sbjct: 20 LSRAGKLNEAKEFIESATVDHGLCL--WRILLGACKNHRNYELGVYAGEKLVELGSPESS 193
Query: 545 GYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGSSWIEIRNVVTAFVSGNNSSPYMAD 604
YV+LS++Y A G + E VR+ M +GV K PG SWIE++ +V FV G+N P + +
Sbjct: 194 AYVLLSSIYTALGDRENVERVRRIMKARGVNKEPGCSWIELKGLVHVFVVGDNQHPQVDE 373
Query: 605 ISNILYFLEIEM 616
I LE+E+
Sbjct: 374 IR-----LELEL 394
>BF646834 weakly similar to PIR|A84784|A847 hypothetical protein At2g36730
[imported] - Arabidopsis thaliana, partial (10%)
Length = 602
Score = 73.9 bits (180), Expect = 2e-13
Identities = 46/135 (34%), Positives = 70/135 (51%), Gaps = 1/135 (0%)
Frame = +2
Query: 17 SLARSGRICHARKLFDEMPDRDS-VAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSF 75
SL+ + HARKL + S ++WN +I Y+ +S+ +F MR + KP+
Sbjct: 194 SLSPFKNLSHARKLVFHFSNNPSPISWNILIRGYASSDSPIESIWVFKKMRENGVKPNKL 373
Query: 76 SYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEM 135
+Y +CA G +HA +V G S + V N++I+ YG C K ARKVFDEM
Sbjct: 374 TYPFIFKSCAMALVLCEGKQVHADLVKFGLDSDVYVCNNMINFYGXCKKIVYARKVFDEM 553
Query: 136 ADSNEVTWCSLLFAY 150
V+W S++ A+
Sbjct: 554 CVXTIVSWNSVMTAW 598
Score = 68.6 bits (166), Expect = 7e-12
Identities = 45/191 (23%), Positives = 86/191 (44%), Gaps = 2/191 (1%)
Frame = +2
Query: 226 CMVHGFVIKSGWSSAMEVKNSILSFYA--KLECPSDAMEMFNSFGAFNQVSWNAIIDAHM 283
CMV F + + + NS+ S +L+ + N +Q+ + +
Sbjct: 35 CMVR-FQTLTNKQQCLNLLNSLHSITKLHQLQAQIHLNSLHNDTHILSQLVYFFSLSPFK 211
Query: 284 KLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGA 343
L +K F P +SW +I GY + + ++ +F M N ++ + L
Sbjct: 212 NLSHARKLVFHFSNNPSP--ISWNILIRGYASSDSPIESIWVFKKMRENGVKPNKLTYPF 385
Query: 344 VLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKD 403
+ +CA +L GK VH+ +++ GLD ++V N+++N Y C + + F +
Sbjct: 386 IFKSCAMALVLCEGKQVHADLVKFGLDSDVYVCNNMINFYGXCKKIVYARKVFDEMCVXT 565
Query: 404 LVSWNSMLFAF 414
+VSWNS++ A+
Sbjct: 566 IVSWNSVMTAW 598
Score = 57.4 bits (137), Expect = 2e-08
Identities = 30/110 (27%), Positives = 59/110 (53%)
Frame = +2
Query: 172 IAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVHGF 231
I+WN +I G+A + +FK+M E+ +P++ T+ + +C + + G VH
Sbjct: 266 ISWNILIRGYASSDSPIESIWVFKKMRENGVKPNKLTYPFIFKSCAMALVLCEGKQVHAD 445
Query: 232 VIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDA 281
++K G S + V N++++FY + A ++F+ VSWN+++ A
Sbjct: 446 LVKFGLDSDVYVCNNMINFYGXCKKIVYARKVFDEMCVXTIVSWNSVMTA 595
Score = 34.7 bits (78), Expect = 0.11
Identities = 14/54 (25%), Positives = 33/54 (60%)
Frame = +2
Query: 405 VSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEG 458
+SWN ++ + E++ +F++M +GVKP+++T+ + +C+ ++ EG
Sbjct: 266 ISWNILIRGYASSDSPIESIWVFKKMRENGVKPNKLTYPFIFKSCAMALVLCEG 427
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,134,791
Number of Sequences: 36976
Number of extensions: 261434
Number of successful extensions: 1411
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 1159
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1352
length of query: 630
length of database: 9,014,727
effective HSP length: 102
effective length of query: 528
effective length of database: 5,243,175
effective search space: 2768396400
effective search space used: 2768396400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0238.21


