

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
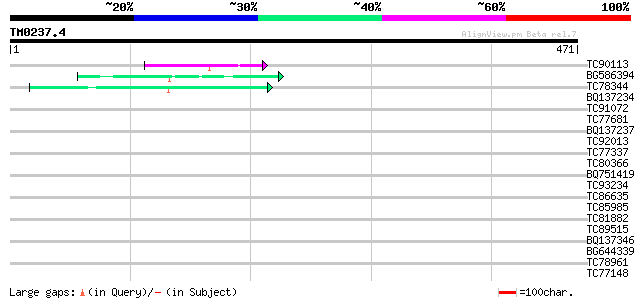
Query= TM0237.4
(471 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protei... 43 3e-04
BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophi... 42 4e-04
TC78344 similar to GP|13540405|gb|AAK29456.1 histone H1 {Lens cu... 42 7e-04
BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkho... 39 0.003
TC91072 weakly similar to GP|22136510|gb|AAM91333.1 unknown prot... 39 0.004
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 39 0.004
BQ137237 weakly similar to GP|11993889|gb| virion-associated nuc... 39 0.006
TC92013 homologue to GP|11595557|emb|CAC18142. related to c-modu... 39 0.006
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 38 0.007
TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like... 38 0.007
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 37 0.013
TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed prot... 37 0.013
TC86635 weakly similar to GP|9758600|dbj|BAB09233.1 gene_id:MDF2... 37 0.013
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 37 0.016
TC81882 similar to GP|9759349|dbj|BAB10004.1 contains similarity... 37 0.021
TC89515 weakly similar to GP|11994144|dbj|BAB01165. contains sim... 36 0.028
BQ137346 homologue to GP|15027095|em hypothetical protein LT.09 ... 36 0.028
BG644339 weakly similar to PIR|T48401|T484 histone deacetylase-l... 35 0.048
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 35 0.048
TC77148 ENOD20 35 0.048
>TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protein
[imported] - Arabidopsis thaliana, partial (70%)
Length = 1259
Score = 42.7 bits (99), Expect = 3e-04
Identities = 31/113 (27%), Positives = 46/113 (40%), Gaps = 11/113 (9%)
Frame = +1
Query: 113 PSRENSPAPNPAK-SKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHR------- 164
P R SP P P + S PP+ + + ++ S S RR RS+S SP R
Sbjct: 886 PGRRRSPPPPPRRRSPPRRARSPPRRSPVGRRRSRSPIRRSARSRSRSFSPRRGRPPMRR 1065
Query: 165 ---SKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQP 214
S S + SP K S +++ P++ A + PPP + P
Sbjct: 1066GRSSSYSDSPSPRKVSRRSKSRSPRRPLK-GRASSNSSSSSSPPPARKPXIIP 1221
Score = 39.3 bits (90), Expect = 0.003
Identities = 26/90 (28%), Positives = 40/90 (43%), Gaps = 4/90 (4%)
Frame = +1
Query: 130 GVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSP----SKESACQETQGA 185
G PP + S + ++S+ R S +SP RS S + SP K SA +G
Sbjct: 4 GYRAPPSSLSSSSRSSSRSRSRSRSFSSSSRSPSRSSKSRSRSPPPPRRKSSAEPARRGR 183
Query: 186 TSPIREAEALPGKDDNPMPPPKSTATVQPS 215
+ P+ P + P PPP+ + V+ S
Sbjct: 184 SPPLPP----PPQSKRPSPPPRKPSPVRES 261
Score = 35.4 bits (80), Expect = 0.048
Identities = 40/139 (28%), Positives = 58/139 (40%), Gaps = 22/139 (15%)
Frame = +1
Query: 98 ATIQADVHEHSISNPPSRENSPAPNPAKSKAE-------GVE------KPPKAASLAKKT 144
A I +V + + PP ++ SP P K + G E K P+ +S +K
Sbjct: 454 AQIDGNVVKARFTLPPRQKASPPPKAVAPKRDAPRTDNAGAEVEKDGPKRPRESSPRRKP 633
Query: 145 STSEGRRK-VRSKSGHKSPHRSKSSTNSSPSK-----ESACQETQGATSPIREAEALPGK 198
S RR V ++G SP R +S + S +S + G T P R PG+
Sbjct: 634 PPSPRRRSPVPRRAG--SPRRPESPRRRADSPVRRRLDSPYRRGAGDTPP-RRRPVSPGR 804
Query: 199 DDNPMPPP---KSTATVQP 214
+P PPP +S A V P
Sbjct: 805 GRSPSPPPRRLRSPARVSP 861
>BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 42.4 bits (98), Expect = 4e-04
Identities = 45/179 (25%), Positives = 66/179 (36%), Gaps = 8/179 (4%)
Frame = +1
Query: 57 KPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGK-EGDGDATIQADVHEHSISNPPSR 115
KP + K K +TT +PE N + + D E + + P S
Sbjct: 247 KPEKETTTKTTKPEAETTT----------EPETNTPTKSETDVPETTTTLEKNQNVPGST 396
Query: 116 ENSPAPNPAKSKAEGV-------EKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSS 168
S P KS+ E EKP + K T+ SE + ++ P S+ +
Sbjct: 397 TESETETPTKSETETTTTEETPTEKPE--TDIPKTTTASEKNKTTTTEP--TVPTVSEET 564
Query: 169 TNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVS 227
P KE+ T+P EA P D+ P P K+ AT P + +NVS
Sbjct: 565 PTEKPEKETT-------TTPETEATTQPETDNTPTNPEKNKATTTPETETTTASETNVS 720
>TC78344 similar to GP|13540405|gb|AAK29456.1 histone H1 {Lens culinaris},
partial (86%)
Length = 1215
Score = 41.6 bits (96), Expect = 7e-04
Identities = 43/211 (20%), Positives = 84/211 (39%), Gaps = 9/211 (4%)
Frame = +2
Query: 17 ISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTS 76
+S V + ++P ++ + + A K K ++PKP AS+ T
Sbjct: 107 VSAVEQTIVEEPAAVDPLPPVVNESDEPTAAKPKKAPKEPKPRKPASKN------TRTHP 268
Query: 77 GASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEG------ 130
++ +DA + K G + + E + P + + K A G
Sbjct: 269 TYEEMVTDAIVTLKEKNGSSQYALAKFIEEKHKNLPANFKKILLVQIKKLVASGKLVKVK 448
Query: 131 --VEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSP 188
+ P K+A+ AKK + ++ + K ++K+ +KS ++P+K A + + A +
Sbjct: 449 GSYKLPAKSAAPAKKPAAAKPKPKPKAKAPVAKAPAAKSKAKAAPAKAKAKAKAKAAPAK 628
Query: 189 IR-EAEALPGKDDNPMPPPKSTATVQPSPQA 218
+ A+A P P K A +P +A
Sbjct: 629 AKPAAKAKPAAKAKPAAKAKPVAKAKPKAKA 721
>BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkholderia
cepacia}, partial (6%)
Length = 1184
Score = 39.3 bits (90), Expect = 0.003
Identities = 35/152 (23%), Positives = 58/152 (38%), Gaps = 9/152 (5%)
Frame = +3
Query: 40 DDNVSLADKLKGRKRKPKPAASAS---------QKRAKGAGVSTTSGASKLASDAQPEVN 90
+DN KG K + A + + +KRA+ AG + GA + QPE
Sbjct: 66 EDNKHEISPTKGNKNRSTTAGAGALQPTDPPGCKKRARRAGTAR-HGAGR-GPPTQPEGR 239
Query: 91 GKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGR 150
GK+G + + RE + N AK G +K K EGR
Sbjct: 240 GKKGREKQEAEPEAKRKK------RERTKTKNKAKKGRRGNKKEKKTTPKETGRERREGR 401
Query: 151 RKVRSKSGHKSPHRSKSSTNSSPSKESACQET 182
R+ ++ PH ++ ++ +E ++T
Sbjct: 402 RRRSTEERRARPHEQEAEKTNAQRRERQKEQT 497
Score = 30.4 bits (67), Expect = 1.5
Identities = 27/119 (22%), Positives = 42/119 (34%), Gaps = 6/119 (5%)
Frame = +2
Query: 90 NGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPP------KAASLAKK 143
N +GDG A RE PA K E +K + A+
Sbjct: 359 NNPKGDG-----AREKRREKEKKHGREKGPAARTGSRKDERTKKRAAKRANNNREARAQH 523
Query: 144 TSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNP 202
T+ R + R ++ + PH S ++ ++ E + +P E E G DNP
Sbjct: 524 TTHRNARPQERRRASEQRPHESPATPVTTARARQKESEKRQGDAPPSEDETRRGTTDNP 700
>TC91072 weakly similar to GP|22136510|gb|AAM91333.1 unknown protein
{Arabidopsis thaliana}, partial (15%)
Length = 1021
Score = 38.9 bits (89), Expect = 0.004
Identities = 50/223 (22%), Positives = 88/223 (39%), Gaps = 24/223 (10%)
Frame = +2
Query: 7 EVITQPSPQVISPVAHSSKDKPVVIEDDDDDD--EDDNVSLADKLKG----RKRK-PKPA 59
++I P I ++ + EDDDDD+ +D V++ KL G +KRK K
Sbjct: 350 QLIELPKLLAIEGFQEKREEYETISEDDDDDNDQSEDVVNVESKLDGNATSKKRKASKKL 529
Query: 60 ASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGD----GDATIQADVHEHSI----SN 111
S S+K+ K +STT + + +A E NG D + + D+ + S N
Sbjct: 530 KSPSKKKIK---MSTTENLADI-PEAHEEENGSVKDCTHRKASLAKKDIGKSSTLSCQKN 697
Query: 112 PPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSK----- 166
S S + + EG K + S +K + + R+++ K K +
Sbjct: 698 DKSGSTSEETRSLQPQDEGEPKKLPSRSARRKKAKRKWLRELKLKEKEKEKEKENENEKD 877
Query: 167 ----SSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPP 205
S ++ ++ G S +R+ + D+ M P
Sbjct: 878 KLHPSQVLEKDDQQIPIKDNNGKVSDVRQQSNEESEADDDMVP 1006
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 38.9 bits (89), Expect = 0.004
Identities = 44/203 (21%), Positives = 80/203 (38%), Gaps = 1/203 (0%)
Frame = +2
Query: 32 EDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPE-VN 90
E+ + D ED+++ L + K +PK A K A + A A+ + V+
Sbjct: 386 EEGETDSEDEDIPLITTEQNGKPEPK-AEELKVSEPKKADAKNAAPAKNAATAKHVKVVD 562
Query: 91 GKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGR 150
K+ D D ++D S + +S + + S + E+ P KK
Sbjct: 563 PKDEDSDDDDESDDEIGSSDDEMENADSDSEDEDDSDEDDEEETP-----VKKVDQG--- 718
Query: 151 RKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTA 210
+K ++S K+P SK S N++P K + AT ++ P D
Sbjct: 719 KKRPNESASKTPISSKKSKNATPEKTDGKKAGHTATPHPKKGGKTPNSD----------- 865
Query: 211 TVQPSPQAKGGFSSNVSSEKLDT 233
+P++ GG S + S+ ++
Sbjct: 866 --AKTPKSGGGLSCSSCSKTFNS 928
Score = 34.3 bits (77), Expect = 0.11
Identities = 31/103 (30%), Positives = 44/103 (42%), Gaps = 5/103 (4%)
Frame = +2
Query: 32 EDDDDDDEDDNVSL-ADKLKGRKRKPKPAAS---ASQKRAKGAGVSTTSG-ASKLASDAQ 86
ED+DD DEDD K+ K++P +AS S K++K A T G + +
Sbjct: 653 EDEDDSDEDDEEETPVKKVDQGKKRPNESASKTPISSKKSKNATPEKTDGKKAGHTATPH 832
Query: 87 PEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAE 129
P+ GK + DA S S+ NS SKA+
Sbjct: 833 PKKGGKTPNSDAKTPKSGGGLSCSSCSKTFNSETGLTQHSKAK 961
>BQ137237 weakly similar to GP|11993889|gb| virion-associated
nuclear-shuttling protein {Mus musculus}, partial (4%)
Length = 1215
Score = 38.5 bits (88), Expect = 0.006
Identities = 41/199 (20%), Positives = 69/199 (34%), Gaps = 7/199 (3%)
Frame = +2
Query: 52 RKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISN 111
RK KP+P + +Q+RAKGA +K KEG G +
Sbjct: 224 RKHKPRPTPTQTQRRAKGAPKEQGRQTTKKEKTRTRAGREKEGGG-------AKRTGTPD 382
Query: 112 PPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNS 171
P ++ + G E+ + A+ A+ ++G R+ RS + H T +
Sbjct: 383 PGQKDRRATETGTEEARAGEEEDRRKATRAR---GNDGGRRKRSDAKRADAHDHNPRTRA 553
Query: 172 SPSKESACQE---TQGATSPIREAEALPGKDDNPMPPPKSTAT----VQPSPQAKGGFSS 224
+ + ++ T+ T P DD PK+ Q P + SS
Sbjct: 554 TANHKTTAPHNTPTESHTQHRGRTPTSPAYDDRRCREPKNAKKQRT*AQQRPAGRAARSS 733
Query: 225 NVSSEKLDTLFEEDPLAAL 243
D + P ++L
Sbjct: 734 TARGHAQDKRHDTRPDSSL 790
>TC92013 homologue to GP|11595557|emb|CAC18142. related to c-module-binding
factor {Neurospora crassa}, partial (2%)
Length = 1437
Score = 38.5 bits (88), Expect = 0.006
Identities = 38/176 (21%), Positives = 62/176 (34%), Gaps = 12/176 (6%)
Frame = -2
Query: 51 GRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSIS 110
G K P PAA SQ + A + K+ S + K+ AT ++S
Sbjct: 899 GDKSPPPPAAGTSQLTSMAARAAARGRKFKITSRKSRGSSAKDATPSAT-------PALS 741
Query: 111 NPPSRENSPAPNPAKS---------KAEGVEKPPKAASLAKKTST---SEGRRKVRSKSG 158
P++E+S PN ++ +E A S K S +E ++
Sbjct: 740 EMPTKESSKEPNNEQAVLPTTEMHDSSEEATASAPAVSPVKSPSPPRPTEAAKEATPDLS 561
Query: 159 HKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQP 214
+P S+S ++E +S +PG P PP S+ +P
Sbjct: 560 PPAPRASRSLHRDDANEEEVIAPAASPSSVSSSDSGIPGASPPPPPPTSSSRRGRP 393
Score = 29.3 bits (64), Expect = 3.4
Identities = 27/116 (23%), Positives = 44/116 (37%), Gaps = 11/116 (9%)
Frame = -2
Query: 112 PPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTS-----TSEGR------RKVRSKSGHK 160
PP + + P A A PP AA ++ TS + GR RK R S
Sbjct: 950 PPKNKPAVPPTVAVEDAGDKSPPPPAAGTSQLTSMAARAAARGRKFKITSRKSRGSSAKD 771
Query: 161 SPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSP 216
+ + + + P+KES+ +E + + E ++ P + PSP
Sbjct: 770 ATPSATPALSEMPTKESS-KEPNNEQAVLPTTEMHDSSEEATASAPAVSPVKSPSP 606
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 38.1 bits (87), Expect = 0.007
Identities = 33/108 (30%), Positives = 46/108 (42%), Gaps = 12/108 (11%)
Frame = -2
Query: 136 KAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKES-ACQETQGATSPIREAEA 194
KA+S + +S+S S S P S SS++SS S S + + ++SP A
Sbjct: 751 KASSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSPFSSAPP 572
Query: 195 LPGK----------DDNPMPPPKSTA-TVQPSPQAKGGFSSNVSSEKL 231
P + P PPP S + PSP A SS+ SS L
Sbjct: 571 APPSGAFFELLLLLELLPYPPPSSDDNSSSPSPNAPSPSSSSSSSSSL 428
>TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 814
Score = 38.1 bits (87), Expect = 0.007
Identities = 29/116 (25%), Positives = 44/116 (37%), Gaps = 1/116 (0%)
Frame = +3
Query: 110 SNPPSRENSPAPN-PAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSS 168
S PP+ +SP P+ PA PP + T + + P +S
Sbjct: 6 SPPPATPSSPPPSTPANPPPVTPSAPPPS------TPATPSSPPPSTTPSAPPPSTPSNS 167
Query: 169 TNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSS 224
S P+ + + G T+P + P DD+P PP T PSP + S+
Sbjct: 168 PPSPPTTPAISPPSGGGTTPSPPSRTPPSSDDSPSPPSSKTPP-PPSPPSSSSIST 332
Score = 28.1 bits (61), Expect = 7.6
Identities = 44/186 (23%), Positives = 64/186 (33%), Gaps = 46/186 (24%)
Frame = +3
Query: 112 PPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEG-------------------RRK 152
PPS ++SP+P SK PP ++S++ T T G R+K
Sbjct: 246 PPSSDDSPSP--PSSKTPPPPSPPSSSSIS--TGTVIGIAVGAVVVLVFFSICCICFRKK 413
Query: 153 VRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKD---DNPMP----P 205
R + + + + + P + E G + A P D P P P
Sbjct: 414 KRRRRDEE--YYGQQNYQQPPPAQRPKVEPYGGPPQQWQNNAHPPSDHVVSKPPPPAPIP 587
Query: 206 PKSTATVQPSP------QAKGGFSSNVSSEKL--------------DTLFEEDPLAALDG 245
P+ + V P P + GG SN S +L T E+ A DG
Sbjct: 588 PRPPSHVAPPPPPPAFISSSGGSGSNYSGGELLPPPSPGIAFSSGKSTFTYEELARATDG 767
Query: 246 FLDGTL 251
F D L
Sbjct: 768 FSDANL 785
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 37.4 bits (85), Expect = 0.013
Identities = 40/179 (22%), Positives = 67/179 (37%), Gaps = 20/179 (11%)
Frame = +1
Query: 56 PKPAAS----------ASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVH 105
P+PAAS +S A+ + TT+ ++ + + + A
Sbjct: 7 PQPAASNLHTTWQPPASSSPSARPSPPPTTASSTPPGAPTPSPTPTRRATLNGNTPAPAS 186
Query: 106 EHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKS---- 161
+ + PP+ ++ AP+P+ G +AS TS + S S H S
Sbjct: 187 PTAQATPPTGPSTAAPSPSTCTTTGPTSSSTSASSPAATSP-----RTTSPSPHSSGTPP 351
Query: 162 ---PHRSKSSTNSSP---SKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQP 214
P S+SST+ P ++ +A + + AT P R P PPP +P
Sbjct: 352 APAPSASRSSTSRPPRARARRAASRSSPWAT-PARACTTAPTSASRQTPPPSPRTAAKP 525
Score = 31.2 bits (69), Expect = 0.90
Identities = 31/129 (24%), Positives = 51/129 (39%), Gaps = 10/129 (7%)
Frame = +1
Query: 110 SNPPSRENSPAPNPAKSKAEGVEKPP----KAASL-----AKKTSTSEGRRKVRSKSGHK 160
S+ PS SP P A S G P + A+L A + T++ +
Sbjct: 55 SSSPSARPSPPPTTASSTPPGAPTPSPTPTRRATLNGNTPAPASPTAQATPPTGPSTAAP 234
Query: 161 SPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPK-STATVQPSPQAK 219
SP + T + P+ S + ATSP + + P P P S ++ P+A+
Sbjct: 235 SP---STCTTTGPTSSSTSASSPAATSPRTTSPSPHSSGTPPAPAPSASRSSTSRPPRAR 405
Query: 220 GGFSSNVSS 228
+++ SS
Sbjct: 406 ARRAASRSS 432
>TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed protein product
{Homo sapiens}, partial (11%)
Length = 591
Score = 37.4 bits (85), Expect = 0.013
Identities = 37/155 (23%), Positives = 62/155 (39%), Gaps = 3/155 (1%)
Frame = +2
Query: 54 RKPKPAASASQKRAKGAGVS---TTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSIS 110
+ PK + ++ QK ++ + S T + AS +S + P + ++ H + +
Sbjct: 86 KSPKSSRNSQQKWSRTS*SSERATPASASLTSSSSIPSQRSRTSRSPSSRPR--HTSTGT 259
Query: 111 NPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTN 170
PP +SPA +P + P + S + STS R S + +P RS+ +
Sbjct: 260 APPFAVSSPASSPTTPSS--TRSSPGSTSTPRTPSTSFSARPPPS-TPPPTPFRSRPTKG 430
Query: 171 SSPSKESACQETQGATSPIREAEALPGKDDNPMPP 205
SSPS + TSP P PP
Sbjct: 431 SSPSSTPSSSSPPAPTSPAISPSRPSAPTRRPSPP 535
Score = 30.8 bits (68), Expect = 1.2
Identities = 28/102 (27%), Positives = 39/102 (37%), Gaps = 12/102 (11%)
Frame = +2
Query: 135 PKAASLAKKTSTSEGRRKVRSKSGHKSPHRSK--------SSTNSSPSKESACQETQGAT 186
P +ASL +S R + + H S SS SSP+ S+ + + G+T
Sbjct: 158 PASASLTSSSSIPSQRSRTSRSPSSRPRHTSTGTAPPFAVSSPASSPTTPSSTRSSPGST 337
Query: 187 S----PIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSS 224
S P A P P P +S T SP + SS
Sbjct: 338 STPRTPSTSFSARPPPSTPPPTPFRSRPTKGSSPSSTPSSSS 463
>TC86635 weakly similar to GP|9758600|dbj|BAB09233.1
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1963
Score = 37.4 bits (85), Expect = 0.013
Identities = 39/153 (25%), Positives = 65/153 (41%)
Frame = +2
Query: 32 EDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNG 91
E++DD+ E+D + +K + + S ++ V SK +S A+ E G
Sbjct: 929 EENDDEPEND---IPEKSEDETPQKSEREDKSDSGSESEDVKKRKRPSKTSS-AKKESAG 1096
Query: 92 KEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRR 151
+ T ++ V S S PP R + N + + + PK S KK S G++
Sbjct: 1097 RS----KTEKSTVTNKSRSPPPKRAPKKSSNTRPKSDDDINESPKVFS-RKKKSEQGGKQ 1261
Query: 152 KVRSKSGHKSPHRSKSSTNSSPSKESACQETQG 184
K+ + + KS SS SKE + T+G
Sbjct: 1262 KISTPTPTKS---------SSKSKEKTEKVTKG 1333
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 37.0 bits (84), Expect = 0.016
Identities = 42/145 (28%), Positives = 58/145 (39%), Gaps = 2/145 (1%)
Frame = +3
Query: 55 KPK-PAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPP 113
KPK PA AS K + A T + + S A P K + + A V P
Sbjct: 198 KPKSPAPVASPKSSPPASSPTAATVTPAVSPAAPVPVAKSPAASSPVVAPVSTPPKPAPV 377
Query: 114 SRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNS-S 172
S +P P + V PP A++ A T ++E SKS K+ K S + S
Sbjct: 378 SSPPAPVPVSSPPTPVPVSSPPTASTPA-VTPSAEVPAAAPSKSKKKTKKGKKHSAPAPS 554
Query: 173 PSKESACQETQGATSPIREAEALPG 197
P+ E GA P +A + PG
Sbjct: 555 PALEGPPAPPVGAPGPSLDASS-PG 626
>TC81882 similar to GP|9759349|dbj|BAB10004.1 contains similarity to unknown
protein~gb|AAF72944.1~gene_id:MAH20.11 {Arabidopsis
thaliana}, partial (5%)
Length = 875
Score = 36.6 bits (83), Expect = 0.021
Identities = 40/184 (21%), Positives = 68/184 (36%), Gaps = 7/184 (3%)
Frame = +2
Query: 12 PSPQVISPVAHSSKDKPVVIEDDDDD-DEDDNVSLADKLKGRKRKPKPAASASQKRAKGA 70
P+ P A K P ++ DD+ D D+ K +PKP++S+S K
Sbjct: 176 PTTVPSKPSAPKPKKPPKLLSFADDEIDADNETPRPRSSKPHHHRPKPSSSSSHKITTHK 355
Query: 71 GVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKA-- 128
T+ S S+ QP+ + +Q + SR S P P+
Sbjct: 356 NRITSHSPSPSPSNVQPQAGTYTLEALRELQKNTRTLVTPTTASRPISSEPKPSSEPVIG 535
Query: 129 -EGVEKPPKAASLAKKTSTSEGRRKVRS---KSGHKSPHRSKSSTNSSPSKESACQETQG 184
+G+ KP + + E K S K+G S + ++ +K ++ G
Sbjct: 536 LKGLLKPVTSEPESDSEENGEFEAKFASVGIKNGKDSFFPGEEDIKAAKAKRERMRKA-G 712
Query: 185 ATSP 188
A +P
Sbjct: 713 AAAP 724
Score = 33.5 bits (75), Expect = 0.18
Identities = 26/117 (22%), Positives = 42/117 (35%), Gaps = 13/117 (11%)
Frame = +2
Query: 114 SRENSPAPNPAKSKAEGVEKPPKAASLA-------------KKTSTSEGRRKVRSKSGHK 160
S +++P P+K A +KPPK S A + + R K S S HK
Sbjct: 161 SDDDTPTTVPSKPSAPKPKKPPKLLSFADDEIDADNETPRPRSSKPHHHRPKPSSSSSHK 340
Query: 161 SPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQ 217
T+ SPS + + Q T + L + P ++ + P+
Sbjct: 341 ITTHKNRITSHSPSPSPSNVQPQAGTYTLEALRELQKNTRTLVTPTTASRPISSEPK 511
>TC89515 weakly similar to GP|11994144|dbj|BAB01165. contains similarity to
nodulin~gene_id:K10D20.11 {Arabidopsis thaliana},
partial (43%)
Length = 854
Score = 36.2 bits (82), Expect = 0.028
Identities = 20/69 (28%), Positives = 34/69 (48%)
Frame = +2
Query: 102 ADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKS 161
++ ++ S + PPS ++S P+PA S EG PP T+TS+ H
Sbjct: 467 SNTNQTSNATPPSPQSSSPPSPAPSNQEGQSPPPDTNQTPPPTATSD----------HDH 616
Query: 162 PHRSKSSTN 170
P R+ +++N
Sbjct: 617 PPRNGAASN 643
>BQ137346 homologue to GP|15027095|em hypothetical protein LT.09 {Leishmania
major}, partial (0%)
Length = 1101
Score = 36.2 bits (82), Expect = 0.028
Identities = 36/160 (22%), Positives = 54/160 (33%), Gaps = 9/160 (5%)
Frame = +1
Query: 52 RKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDA---------TIQA 102
R + P A A+ + +GA +G + A P N + G T +
Sbjct: 88 RNKSGSPTAEAAAQPTRGAPGRQETGTRR-AGQTPPRTNQRNHSGQGQSRSAKTPRTHTS 264
Query: 103 DVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSP 162
D H+ S P+R N+P K+ + KT E RK
Sbjct: 265 DKHQPQTSFRPARHNTPPHGTRKTTPRRRRNKQRTTGKQPKTKAGENARK---------- 414
Query: 163 HRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNP 202
+K PS E + +GA + R GK +NP
Sbjct: 415 --TKEGQTGEPSGE---KPDKGARNRDRPNRGTTGKRNNP 519
>BG644339 weakly similar to PIR|T48401|T484 histone deacetylase-like protein
- Arabidopsis thaliana, partial (26%)
Length = 763
Score = 35.4 bits (80), Expect = 0.048
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Frame = +1
Query: 32 EDDDDDDEDDNVSLADKLKGRKRKPKPA---ASASQKRAKGAGVSTTSGASK 80
EDDDD +EDD+ K +K++P P+ A S K+AK + SG K
Sbjct: 583 EDDDDSEEDDSEEETPKKVEQKKRPAPSPKVAPGSGKKAKQVTPAHKSGGKK 738
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 35.4 bits (80), Expect = 0.048
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 2/100 (2%)
Frame = +2
Query: 134 PPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTN--SSPSKESACQETQGATSPIRE 191
P K + A+ S S GR RS + +SP RS+S S S + + SP+R
Sbjct: 476 PKKLSRHARSVSRSPGRS--RSPARSRSPARSRSPRRGRSRDRSYSPARSYSRSRSPVRR 649
Query: 192 AEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEKL 231
+ D + PPP + SP+ S++ ++++
Sbjct: 650 DRSPVASDRSRSPPPSKSRKYSRSPEGSPQKSTSPGNDRV 769
>TC77148 ENOD20
Length = 1108
Score = 35.4 bits (80), Expect = 0.048
Identities = 23/84 (27%), Positives = 40/84 (47%), Gaps = 3/84 (3%)
Frame = +3
Query: 108 SISNPPSRENSPAPNPAKS---KAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHR 164
S+ +PPS SP+P+P+ S ++ + P K + + S S + S+S +P
Sbjct: 492 SLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSP 671
Query: 165 SKSSTNSSPSKESACQETQGATSP 188
S S + +PS + + A SP
Sbjct: 672 SDSVASLAPSSSPSDESPSPAPSP 743
Score = 32.0 bits (71), Expect = 0.53
Identities = 31/135 (22%), Positives = 49/135 (35%), Gaps = 1/135 (0%)
Frame = +3
Query: 161 SPHRSKSSTN-SSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAK 219
SP +SST P + S + SP P P+P P+ + PSP
Sbjct: 441 SPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPS 620
Query: 220 GGFSSNVSSEKLDTLFEEDPLAALDGFLDGTLDWDSPPHQADATAETAQSSGVANLQQIE 279
S + S D +A+L + D P A + + + G A +E
Sbjct: 621 LSKSPSPSESPSLAPSPSDSVASL---APSSSPSDESPSPAPSPSSSGSKGGGAGHGFLE 791
Query: 280 ESMGRLKAIVFDIGF 294
S+ + ++F I F
Sbjct: 792 VSIAMMMFLIF*IMF 836
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.307 0.125 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,515,411
Number of Sequences: 36976
Number of extensions: 142445
Number of successful extensions: 2211
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 1520
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1871
length of query: 471
length of database: 9,014,727
effective HSP length: 100
effective length of query: 371
effective length of database: 5,317,127
effective search space: 1972654117
effective search space used: 1972654117
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0237.4


