

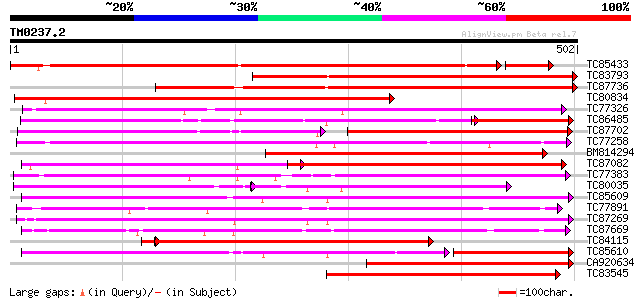
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0237.2
(502 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85433 weakly similar to GP|6002279|emb|CAB56741.1 cytochrome P... 415 e-128
TC83793 weakly similar to GP|6002279|emb|CAB56741.1 cytochrome P... 446 e-125
TC87736 weakly similar to GP|6002279|emb|CAB56741.1 cytochrome P... 424 e-119
TC80834 similar to GP|6002279|emb|CAB56741.1 cytochrome P450 mon... 360 e-100
TC77326 similar to GP|12004682|gb|AAG44132.1 cytochrome P450 {Pi... 319 2e-87
TC86485 similar to GP|23315211|gb|AAN20392.1 Sequence 405 from p... 244 5e-85
TC87702 weakly similar to SP|O48923|C7DA_SOYBN Cytochrome P450 7... 186 5e-78
TC77258 similar to GP|5059126|gb|AAD38930.1| cytochrome P450 mon... 282 2e-76
BM814294 weakly similar to GP|6002279|emb cytochrome P450 monoox... 280 7e-76
TC87082 weakly similar to GP|7406712|emb|CAB85635.1 putative rip... 280 1e-75
TC77383 similar to SP|O48922|C982_SOYBN Cytochrome P450 98A2 (EC... 276 1e-74
TC80035 weakly similar to GP|18252325|gb|AAL66194.1 cytochrome P... 172 1e-72
TC85609 weakly similar to SP|Q9LTL2|C72P_ARATH Cytochrome P450 7... 269 2e-72
TC77891 weakly similar to SP|P93147|C81E_GLYEC Cytochrome P450 8... 268 5e-72
TC87269 similar to PIR|T05940|T05940 cytochrome P450 83D1p - soy... 267 6e-72
TC87669 weakly similar to GP|3850630|emb|CAA10067.1 cytochrome P... 265 4e-71
TC84115 weakly similar to GP|17065916|emb|CAC80883. geraniol 10-... 263 1e-70
TC85610 similar to GP|21068674|emb|CAD31843. putative cytochrome... 172 3e-70
CA920634 weakly similar to SP|O64636|C7C1 Cytochrome P450 76C1 (... 257 6e-69
TC83545 weakly similar to GP|7406712|emb|CAB85635.1 putative rip... 256 2e-68
>TC85433 weakly similar to GP|6002279|emb|CAB56741.1 cytochrome P450
monooxygenase {Cicer arietinum}, partial (56%)
Length = 1600
Score = 415 bits (1066), Expect(2) = e-128
Identities = 217/438 (49%), Positives = 300/438 (67%), Gaps = 3/438 (0%)
Frame = +1
Query: 1 MDTISCILLFLLTITFILFLANTI---LKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPH 57
MD S +LFLLT F + + KPN+K LPPGP I+ N+ +L +KP
Sbjct: 64 MDYGSGAMLFLLTCIVTYFFGSFLGRSKKPNYK-----LPPGPSFFTIMSNVVELYNKPQ 228
Query: 58 QTLATLSKTHGPIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRY 117
QTLA +K +GPIM +KL TTI++SSS AK L +D+ ++R +PD +N + +
Sbjct: 229 QTLAEFAKLYGPIMRIKLCTETTIIISSSHMAKEILFTNDSLFTDRSVPDNTTTHNHNNF 408
Query: 118 SLPFIPVSHRWRDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGR 177
SL F+P S W+ LRKIC+N LFS K L+ +++LR K+ +L ND+ KS + GEAV+VGR
Sbjct: 409 SLVFLPFSPLWQHLRKICHNNLFSTKTLDGSQELRRRKLKDLLNDMHKSSITGEAVDVGR 588
Query: 178 LAFKTTINLLSNTIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHGI 237
AFK IN LS T S D + + D E+K ++ +++ VG PN+AD FPVLK+ DP GI
Sbjct: 589 AAFKACINFLSYTFVSQDFVENLDD--EYKNIVSTLLSAVGTPNIADHFPVLKIFDPQGI 762
Query: 238 RRRTGSYFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNGSEMYKDKIERL 297
RR T Y ++ ++++R+K R+ Y +KNDMLDA+LD ++ + +M K +I+ L
Sbjct: 763 RRHTTLYVSKVFYALDIIIDQRMKMRQSEQYVSKNDMLDALLDISKEDNQKMDKRQIKHL 942
Query: 298 SVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQA 357
+DL VAGT+T +E M+E++R+PEVMSKAK ELE+TIG GK +EES I+RLPYL A
Sbjct: 943 LLDLLVAGTETSAYGLERAMSEVVRHPEVMSKAKKELEETIGLGKPIEESDIDRLPYLNA 1122
Query: 358 IVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSPE 417
++KE RLHP P+LLPR+A VD+++ GYTIPKG+QVL+N WA GR ++WD+ +LFSPE
Sbjct: 1123VIKESLRLHPPAPMLLPRKARVDVEIAGYTIPKGAQVLINEWAIGR-TDIWDDAHLFSPE 1299
Query: 418 RFLGSEIDFKGRSFELTP 435
RFLGSEID KGR F+LTP
Sbjct: 1300RFLGSEIDVKGRHFKLTP 1353
Score = 62.8 bits (151), Expect(2) = e-128
Identities = 26/42 (61%), Positives = 36/42 (84%)
Frame = +3
Query: 440 RRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPEDMDMDE 481
RRICPG PLA+R+L LMLG LIN F+W+LE ++P+DM++D+
Sbjct: 1368 RRICPGSPLAVRMLHLMLGSLINSFDWKLENNMEPKDMNLDK 1493
>TC83793 weakly similar to GP|6002279|emb|CAB56741.1 cytochrome P450
monooxygenase {Cicer arietinum}, partial (48%)
Length = 923
Score = 446 bits (1147), Expect = e-125
Identities = 216/287 (75%), Positives = 250/287 (86%)
Frame = +2
Query: 216 EVGRPNLADCFPVLKVVDPHGIRRRTGSYFERLMKIFKGLVEERLKQRKESGYNTKNDML 275
EVGR N+ADCFPVLKV+DP GIRRRTG YF +L+ IF+GLV++RLK R+ +GY K+DML
Sbjct: 2 EVGRANIADCFPVLKVLDPIGIRRRTGEYFGKLLGIFQGLVDQRLKMRELNGYCGKSDML 181
Query: 276 DAMLDNAQHNGSEMYKDKIERLSVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELE 335
DAMLD+ + N EMYKDKIERLSVDLFVAGTDTVTST+EW MAELL NP +MSKAKSEL
Sbjct: 182 DAMLDD-EKNAGEMYKDKIERLSVDLFVAGTDTVTSTLEWAMAELLHNPNIMSKAKSELN 358
Query: 336 KTIGKGKLVEESHIERLPYLQAIVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVL 395
+ IGKG VEES I +LPYLQAI+KE FRLHPAVPLLLPR+AE+DL+++GY +PKG+QVL
Sbjct: 359 QIIGKGNSVEESDIGKLPYLQAIIKETFRLHPAVPLLLPRKAEIDLEINGYKVPKGAQVL 538
Query: 396 VNVWANGRDPNLWDNPNLFSPERFLGSEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFL 455
+NVWA GRD NLW+NPN F PERFLGS+IDFKGR+FELTPFG GRRICPGLPLAIR+LFL
Sbjct: 539 INVWAIGRDSNLWENPNEFLPERFLGSDIDFKGRNFELTPFGGGRRICPGLPLAIRVLFL 718
Query: 456 MLGLLINCFNWELEGGIKPEDMDMDETFGLTLEKTQPVLAVPTKTTN 502
MLGL INCF+WEL GGIKPEDM+MD+ F LTLEK QP+L VP K +N
Sbjct: 719 MLGLFINCFDWELVGGIKPEDMNMDDKFELTLEKAQPLLVVPIKVSN 859
>TC87736 weakly similar to GP|6002279|emb|CAB56741.1 cytochrome P450
monooxygenase {Cicer arietinum}, partial (55%)
Length = 1367
Score = 424 bits (1091), Expect = e-119
Identities = 204/374 (54%), Positives = 284/374 (75%), Gaps = 1/374 (0%)
Frame = +3
Query: 130 DLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTINLLSN 189
++R++C N LFSNK+L+A++ LR KI EL N I + LKGEA+++G+LAFKT+INLLSN
Sbjct: 3 EMRRVCKNKLFSNKSLDASQYLRRGKIDELINYISQCSLKGEAIDMGKLAFKTSINLLSN 182
Query: 190 TIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHGIRRRTGSYFERLM 249
T++SVD + K++++ + + VG PN+AD FP+L+++DP GI+R Y +L
Sbjct: 183 TVFSVDFANN-------KDLVMDMSETVGSPNMADFFPLLRLIDPQGIKRTYVFYVGKLF 341
Query: 250 KIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNGSEMYKDKIERLSVDLFVAGTDTV 309
IF +++++LK R+ G+ T NDMLD++L A+ N E+ ++KI+ L DL V GTDT
Sbjct: 342 GIFDNIIDQKLKLREGDGFVTNNDMLDSLL--AEENKKELDREKIQHLLHDLLVGGTDTT 515
Query: 310 TSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIVKEIFRLHPAV 369
T T+EW MAELL NP VMSK K ELE+TIG G +EES + RLPYLQAI+KE RLHP
Sbjct: 516 TYTLEWAMAELLHNPNVMSKVKKELEETIGIGNPIEESDVTRLPYLQAIIKETLRLHPIA 695
Query: 370 PLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSPERFLGSEIDFKGR 429
PLLLPR+A+ D++++GY IPKG+Q+ VNVWA GRDP +WDNPNLFSPERFLG+++D KG+
Sbjct: 696 PLLLPRKAKEDVEVNGYLIPKGAQIFVNVWAIGRDPKVWDNPNLFSPERFLGTKLDIKGQ 875
Query: 430 SFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPEDMDMDETF-GLTLE 488
+F+LTPFG+GRRICPGLPLA+R+L +MLG L+ F+W+LE G+KPE++DM++ GL L
Sbjct: 876 NFQLTPFGSGRRICPGLPLAMRMLHMMLGSLLISFDWKLENGMKPEEIDMEDAIQGLALR 1055
Query: 489 KTQPVLAVPTKTTN 502
K + + +PTK +N
Sbjct: 1056KCESLRVIPTKISN 1097
>TC80834 similar to GP|6002279|emb|CAB56741.1 cytochrome P450 monooxygenase
{Cicer arietinum}, partial (62%)
Length = 1112
Score = 360 bits (924), Expect = e-100
Identities = 184/340 (54%), Positives = 246/340 (72%), Gaps = 4/340 (1%)
Frame = +3
Query: 5 SCILLFLLTITFILFLANTILKPNHK---KKKLPLPPGPKGLPIVGNLFQLGHKPHQTLA 61
S + ++ TF + L L P +K K LPPGP LPI+GNL +LG+KPH +LA
Sbjct: 51 STLSYLVIIFTFSILLLIKFLTPTNKTNQKNHSKLPPGPSQLPIIGNLLKLGNKPHHSLA 230
Query: 62 TLSKTHGPIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPF 121
LS HGPIMTLKLGQ+TTIV+SS++ AK LQ HD LSNR +PDA+ N D+YSL F
Sbjct: 231 NLSNIHGPIMTLKLGQVTTIVISSADIAKEVLQTHDTLLSNRTVPDALSVLNHDQYSLSF 410
Query: 122 IPVSHRWRDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFK 181
+ VS RWRDLRKICNN LF+NK L++++ LR K+ +L +DIKK EAV++GR+AF
Sbjct: 411 MRVSPRWRDLRKICNNQLFANKTLDSSQALRRRKLQDLLDDIKKCSEIEEAVDIGRVAFM 590
Query: 182 TTINLLSNTIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHGIRRRT 241
TTINLLSNT +S D + SA++AGE+KE++VSI+KEVG PNL+D FP+LKV D GIRRR+
Sbjct: 591 TTINLLSNTFFSADFVHSAEEAGEYKEIVVSILKEVGAPNLSDFFPMLKVFDLQGIRRRS 770
Query: 242 GSYFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNGS-EMYKDKIERLSVD 300
++++ IF+ V ERLK R+ +G +D+LDA+L+ + +G EM KD+IE L ++
Sbjct: 771 VVSVKKVLSIFRSFVGERLKMREGTGSIGNDDVLDALLNISLDDGKIEMDKDEIEHLLLN 950
Query: 301 LFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGK 340
+FVAGTDT T T+EW MAEL+ NPE+M K + ELE+ +GK
Sbjct: 951 IFVAGTDTTTYTLEWAMAELMHNPEIMLKVQKELEQVVGK 1070
>TC77326 similar to GP|12004682|gb|AAG44132.1 cytochrome P450 {Pisum
sativum}, complete
Length = 1864
Score = 319 bits (817), Expect = 2e-87
Identities = 182/492 (36%), Positives = 273/492 (54%), Gaps = 10/492 (2%)
Frame = +1
Query: 12 LTITFILFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPHQTLATLSKTHGPIM 71
L TF++ L + ++ K LPPGPK PI+GNL +G PHQ+L L++ +GPIM
Sbjct: 94 LFATFVILL---LFSRRLRRHKYNLPPGPKPWPIIGNLNLIGSLPHQSLHGLTQKYGPIM 264
Query: 72 TLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFIPVSHRWRDL 131
L G IV ++ E AKS L+ HD L+ R A K + + + WR
Sbjct: 265 HLYFGSKPVIVGATVELAKSFLKTHDATLAGRPKLSAGKYTTYNYSDITWSQYGPYWRQA 444
Query: 132 RKICNNLLFSNKNLNANEQLRC----DKILELFNDIKKSMLKGEAVNVGRLAFKTTINLL 187
R++C LFS K L + E +R D + +LFN K++L + ++ ++N++
Sbjct: 445 RRMCLLELFSAKRLESYEYIRKQEMHDFLHKLFNSKNKTILVKDHLST------LSLNVI 606
Query: 188 SNTIYSVDLIRSADKA----GEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHGIRRRTGS 243
S + + D A EFK+++ + G N+ D P + +D G +R +
Sbjct: 607 SRMVLGKKYLEKTDNAVISPDEFKKMLDELFLLNGILNIGDFIPWIHFLDLQGYVKRMKT 786
Query: 244 YFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNGSEMYKDK--IERLSVDL 301
++ + + ++EE +++RK DM+D +L A+ E+ ++ ++ + DL
Sbjct: 787 LSKKFDRFMEHVLEEHIERRKNVKDYVAKDMVDVLLQLAEDPNLEVKLERHGVKAFTQDL 966
Query: 302 FVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIVKE 361
GT++ TVEW ++EL+R PE+ KA EL++ IGK + VEE I LPY+ AI KE
Sbjct: 967 IAGGTESSAVTVEWAISELVRKPEIFKKATEELDRVIGKDRWVEEKDIANLPYVYAIAKE 1146
Query: 362 IFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSPERFLG 421
RLHP P L+PR+A D + GY IPKG+ VLVN W RD +W+NP F PERFLG
Sbjct: 1147TMRLHPVAPFLVPREAREDCKVDGYDIPKGTIVLVNTWTIARDSEVWENPYEFMPERFLG 1326
Query: 422 SEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPEDMDMDE 481
+ID KG FEL PFGAGRR+CPG PL I+++ L L++ FNW L +K ED++M+E
Sbjct: 1327KDIDVKGHDFELLPFGAGRRMCPGYPLGIKVIQTSLANLLHGFNWTLPNNVKKEDLNMEE 1506
Query: 482 TFGLTLEKTQPV 493
FGL+ K P+
Sbjct: 1507IFGLSTPKKIPL 1542
>TC86485 similar to GP|23315211|gb|AAN20392.1 Sequence 405 from patent US
6410718, partial (90%)
Length = 1927
Score = 244 bits (622), Expect(2) = 5e-85
Identities = 139/419 (33%), Positives = 224/419 (53%), Gaps = 12/419 (2%)
Frame = +3
Query: 10 FLLTITFILFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPHQTLATLSKTHGP 69
FL+ I FI+ L + + K+ PPGP GLPI+GN+ + H+ LA L+K +G
Sbjct: 183 FLMAIMFIVPLILLLGLVSRILKRPRYPPGPIGLPIIGNMLMMDQLTHRGLANLAKKYGG 362
Query: 70 IMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFIPVSHRWR 129
I L++G + + +S ++ A+ LQ+ DN SNR A+K DR + F WR
Sbjct: 363 IFHLRMGFLHMVAISDADAARQVLQVQDNIFSNRPATVAIKYLTYDRADMAFAHYGPFWR 542
Query: 130 DLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTINLLSN 189
+RK+C LFS K+ + + +R D++ + ++ G VN+G L F T N++
Sbjct: 543 QMRKLCVMKLFSRKHAESWQSVR-DEVDYAVRTVSDNI--GNPVNIGELVFNLTKNIIYR 713
Query: 190 TIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHGIRRRTGSYFERLM 249
+ S + EF ++ K G N++D P +DP G+ R + L
Sbjct: 714 AAFGSS---SREGQDEFIGILQEFSKLFGAFNISDFVPCFGAIDPQGLNARLVKARKELD 884
Query: 250 KIFKGLVEERLKQRKESGYNTKNDMLDAML------------DNAQHNGSEMYKDKIERL 297
+++E + Q+K+S + + DM+D +L + +N ++ KD I+ +
Sbjct: 885 SFIDKIIDEHV-QKKKSVVDEETDMVDELLAFYSEEAKLNNESDDLNNSIKLTKDNIKAI 1061
Query: 298 SVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQA 357
+D+ GT+TV S +EW MAEL+++PE + K + EL + +G + VEES E+L YL+
Sbjct: 1062IMDVMFGGTETVASAIEWAMAELMKSPEDLKKVQQELAEVVGLSRQVEESDFEKLTYLKC 1241
Query: 358 IVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSP 416
+KE RLHP +PLLL AE + ++GY IPK ++V++N WA GRD N W+ P F P
Sbjct: 1242ALKETLRLHPPIPLLLHETAE-EATVNGYFIPKQARVMINAWAIGRDANCWEEPQSFKP 1415
Score = 89.4 bits (220), Expect(2) = 5e-85
Identities = 42/91 (46%), Positives = 58/91 (63%), Gaps = 1/91 (1%)
Frame = +1
Query: 410 NPNLFSPERFLGSEI-DFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWEL 468
N + S FL + DFKG +FE PFG+GRR CPG+ L + L L + L++CF WEL
Sbjct: 1396 NLRVLSRHXFLKPGVPDFKGSNFEFIPFGSGRRSCPGMQLGLYALDLAVAHLLHCFTWEL 1575
Query: 469 EGGIKPEDMDMDETFGLTLEKTQPVLAVPTK 499
G+KP +MDM + FGLT + ++A+PTK
Sbjct: 1576 PDGMKPSEMDMSDVFGLTAPRASRLVAIPTK 1668
>TC87702 weakly similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (EC
1.14.-.-). [Soybean] {Glycine max}, partial (51%)
Length = 1732
Score = 186 bits (473), Expect(2) = 5e-78
Identities = 85/199 (42%), Positives = 128/199 (63%)
Frame = +2
Query: 300 DLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIV 359
D+F AG+DT +T+EW M+EL++NP+VM KA++E+ + V+E+ + +L YL++++
Sbjct: 926 DIFGAGSDTTFTTLEWAMSELIKNPQVMKKAQAEVRSVYNEKGYVDEASLHKLKYLKSVI 1105
Query: 360 KEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSPERF 419
E RLH +PLLLPRQ +++GY IP S+V+VN W+ RD W F PERF
Sbjct: 1106 TETLRLHAPIPLLLPRQCSEKCEINGYEIPAKSKVIVNAWSICRDSRYWIEAEKFFPERF 1285
Query: 420 LGSEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPEDMDM 479
+ S +D+KG F+ PFGAGRR+CPG+ I L + L L+ F+W + G +D+DM
Sbjct: 1286 IDSSVDYKGVDFQFIPFGAGRRMCPGMTSGIASLEISLANLLFHFDWRMPNGNNADDLDM 1465
Query: 480 DETFGLTLEKTQPVLAVPT 498
DE+FGL + + + VPT
Sbjct: 1466 DESFGLAVRRKHDLRLVPT 1522
Score = 123 bits (308), Expect(2) = 5e-78
Identities = 79/276 (28%), Positives = 139/276 (49%), Gaps = 4/276 (1%)
Frame = +1
Query: 8 LLFLLTITFILFLA-NTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPHQTLATLSKT 66
+L + F+LF+ I + KK LPPGP+ LP++GN+ QLG PHQ LA L++
Sbjct: 43 ILISTILGFLLFMVIKFIWRSKTKKTTYKLPPGPRKLPLIGNIHQLGTLPHQALAKLAQE 222
Query: 67 HGPIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFIPVSH 126
+G +M ++LG+++ IV+SS E AK ++ HD +NR + + + F P
Sbjct: 223 YGSLMHMQLGELSCIVVSSQEMAKEIMKTHDLNFANRPPLLSAEIVTYGYKGMTFSPHGS 402
Query: 127 RWRDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTINL 186
WR +RKIC L S + + R +++ DI S +G +N+ L
Sbjct: 403 YWRQMRKICTMELLSQNRVESFRLQREEELANFVKDINSS--EGSPINLSEKLDSLAYGL 576
Query: 187 LSNTIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDP-HGIRRRTGSYF 245
S T + + + DK +++++I ++K G +LAD +P ++++ G+R+R
Sbjct: 577 TSRTAFGAN-VEDEDKE-KYRKIIKDVLKVAGGFSLADLYPSIRILQVLTGLRQRIEKLH 750
Query: 246 ERLMKIFKGLVEERLKQRKESGYNTK--NDMLDAML 279
KI + +V K+ E+ + D++D +L
Sbjct: 751 GETDKILENIVRSHRKKNLETKAKEEKWEDLVDVLL 858
>TC77258 similar to GP|5059126|gb|AAD38930.1| cytochrome P450
monooxygenaseCYP93D1 {Glycine max}, partial (95%)
Length = 1823
Score = 282 bits (721), Expect = 2e-76
Identities = 166/503 (33%), Positives = 270/503 (53%), Gaps = 12/503 (2%)
Frame = +1
Query: 7 ILLFLLTITFILFLANTILKPNHKKKKLPLPPGPK-GLPIVGNLFQLGHKPHQTLATLSK 65
++LFLL +F+ + K K LPPGP PI+G+ L H++L LS
Sbjct: 100 VVLFLLWFISSIFIRSLFKKSVCYK----LPPGPPISFPILGHAPYLRSLLHKSLYKLSN 267
Query: 66 THGPIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFIPVS 125
+GP+M + LG +V S++E+AK L+ + +NR I A + FIP
Sbjct: 268 RYGPLMHIMLGSQHVVVASTAESAKQILKTCEESFTNRPIMIASENLTYGAADYFFIPYG 447
Query: 126 HRWRDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTIN 185
+ WR L+K+C L S K L +R D+I + + G+ + + + T N
Sbjct: 448 NYWRFLKKLCMTELLSGKTLEHFVHIREDEIKCFMGTLLEISKNGKPIEMRHELIRHTNN 627
Query: 186 LLSNTIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHGIRRRTGSYF 245
++S D+ G+ ++V+ I + +G NL D ++ +D G +R
Sbjct: 628 IISRMTMGKKSSGMNDEVGQLRKVVREIGELLGAFNLGDIIGFMRPLDLQGFGKRNKDTH 807
Query: 246 ERLMKIFKGLVEERLKQRKESGYNT--KNDMLDAMLDNAQHNG---SEMYKDKIERLSVD 300
++ + + +++E + R + G + K D+ D +L+ + + S++ + + ++D
Sbjct: 808 HKMDVMMEKVLKEHEEARAKEGAGSDRKKDLFDILLNLIEADDGAESKLTRQSAKAFALD 987
Query: 301 LFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIVK 360
+F+AGT+ S +EW +AEL+RNP V KA+ E++ T+GK +L +ES I LPYLQA+VK
Sbjct: 988 MFIAGTNGPASVLEWALAELIRNPHVFKKAREEIDSTVGKERLFKESDIPNLPYLQAVVK 1167
Query: 361 EIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSPERFL 420
E R+HP P + R+A + GY +P S++ +N WA GRDPN WDNP +F+PERFL
Sbjct: 1168ETLRMHPPTP-IFAREATRSCQVDGYDVPAFSKIFINAWAIGRDPNYWDNPLVFNPERFL 1344
Query: 421 GSE------IDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKP 474
S+ ID +G+ ++L PFG+GRR CPG LA+ ++ L LI CF+W + G K
Sbjct: 1345QSDDPSKSKIDVRGQYYQLLPFGSGRRSCPGSSLALLVIQATLASLIQCFDWVVNDG-KS 1521
Query: 475 EDMDMDETFGLTLEKTQPVLAVP 497
D+DM E +T+ +P+ P
Sbjct: 1522HDIDMSEVGRVTVFLAKPLKCKP 1590
>BM814294 weakly similar to GP|6002279|emb cytochrome P450 monooxygenase
{Cicer arietinum}, partial (37%)
Length = 793
Score = 280 bits (717), Expect = 7e-76
Identities = 135/251 (53%), Positives = 183/251 (72%), Gaps = 1/251 (0%)
Frame = +1
Query: 227 PVLKVVDPHGIRRRTGSYFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNG 286
P+ +++DP GI+ Y +L +F ++++RLK R+E G+ T NDMLD++LD + N
Sbjct: 1 PLNRLIDPQGIK*TYMFYIGKLFNVFDNIIDQRLKLREEDGFFTNNDMLDSLLDIPEENR 180
Query: 287 SEMYKDKIERLSVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEE 346
E+ ++KIE L DL V GTDT T T+EW MAELL NP +MSK K ELE TIG G +EE
Sbjct: 181 KELDREKIEHLLHDLLVGGTDTTTYTLEWAMAELLHNPNIMSKVKKELEDTIGIGNPLEE 360
Query: 347 SHIERLPYLQAIVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPN 406
S I RLPYLQA++KE RLHP PLLLPR+A+ D++++GYTIPK +Q+ VNVWA GRDP
Sbjct: 361 SDITRLPYLQAVIKETLRLHPIAPLLLPRKAKEDVEVNGYTIPKDAQIFVNVWAIGRDPE 540
Query: 407 LWDNPNLFSPERFLGSEIDFKGRSFELTPFGAGRRICPGLPLAI-RLLFLMLGLLINCFN 465
+WDNP LFSPERFLG+++D KG++F+LTPFG+GRR + +L +MLG L+ F+
Sbjct: 541 VWDNPYLFSPERFLGTKLDIKGQNFQLTPFGSGRRNLSRFGH*L*GMLHMMLGSLLISFD 720
Query: 466 WELEGGIKPED 476
W+LE +KPE+
Sbjct: 721 WKLENDMKPEE 753
>TC87082 weakly similar to GP|7406712|emb|CAB85635.1 putative
ripening-related P-450 enzyme {Vitis vinifera}, partial
(57%)
Length = 1646
Score = 280 bits (715), Expect = 1e-75
Identities = 130/248 (52%), Positives = 186/248 (74%), Gaps = 1/248 (0%)
Frame = +1
Query: 247 RLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLD-NAQHNGSEMYKDKIERLSVDLFVAG 305
+L +IF G++EER++ R ND+LD++L+ N SE+ + ++ L +DLFVAG
Sbjct: 742 KLCEIFDGIIEERIRSRSSKVVEVCNDVLDSLLNINIGEATSELSRSEMVHLFLDLFVAG 921
Query: 306 TDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIVKEIFRL 365
DT +S +EW +AELLRNP+ ++K + EL +TIGKG+ +EESHI +LP+LQA+VKE FRL
Sbjct: 922 IDTTSSIIEWIIAELLRNPDKLTKVRKELCQTIGKGETIEESHIFKLPFLQAVVKETFRL 1101
Query: 366 HPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSPERFLGSEID 425
HP +PLLLP + + +++ G+ +PK +QVLVNVWA GRDP +W NP++F+PERFL +I+
Sbjct: 1102HPPIPLLLPHKCDELVNILGFNVPKNAQVLVNVWAMGRDPTIWKNPDMFAPERFLECDIN 1281
Query: 426 FKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPEDMDMDETFGL 485
+KG +FEL PFGAG+RICPGLPLA R + LM+ L++ F W L G+ PE ++MDE FGL
Sbjct: 1282YKGNNFELIPFGAGKRICPGLPLAHRTMHLMVASLLHNFEWNLADGLIPEQLNMDEQFGL 1461
Query: 486 TLEKTQPV 493
TL++ QP+
Sbjct: 1462TLKRVQPL 1485
Score = 199 bits (506), Expect = 2e-51
Identities = 103/258 (39%), Positives = 156/258 (59%), Gaps = 6/258 (2%)
Frame = +2
Query: 11 LLTITFI--LFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPHQTLATLSKTHG 68
LL I+F+ L + K N + LPPGPK LPI+GN+ +LG PH+ L LSK +G
Sbjct: 17 LLVISFVSATILIFILRKSNQTQNSTKLPPGPKPLPIIGNILELGKNPHKALTKLSKIYG 196
Query: 69 PIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFIPVSHRW 128
PIMTLKLG ITTIV+SS + AK L + SNR +P A+ + D++S+ ++P + W
Sbjct: 197 PIMTLKLGSITTIVISSPQVAKQVLHDNSQIFSNRTVPHAISAVDHDKFSVGWVPTLNLW 376
Query: 129 RDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTINLLS 188
+ LRK C +FS K L++ LR K+ EL + + + KGE ++G F +N +S
Sbjct: 377 KKLRKSCATKVFSTKMLDSTRNLRQQKLQELLDYVNEKSNKGEVFDIGEAVFTNVLNSIS 556
Query: 189 NTIYSVDLIRSA---DKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHG-IRRRTGSY 244
NT++S+DL S +K+ EFK +I IM+E G+PN++D FP+L+ +DP G + + SY
Sbjct: 557 NTLFSMDLAHSTVPDEKSQEFKTIIWGIMEEAGKPNISDFFPILRPLDPQGFVCKDDQSY 736
Query: 245 FERLMKIFKGLVEERLKQ 262
E ++ L+++ Q
Sbjct: 737 EESCVRFLMELLKKEFVQ 790
>TC77383 similar to SP|O48922|C982_SOYBN Cytochrome P450 98A2 (EC 1.14.-.-).
[Soybean] {Glycine max}, complete
Length = 1782
Score = 276 bits (707), Expect = 1e-74
Identities = 168/510 (32%), Positives = 271/510 (52%), Gaps = 17/510 (3%)
Frame = +3
Query: 4 ISCILLFLLTITFI-LFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPHQTLAT 62
I+ L + ++FI +FL T+ + + + LPPGP+ P+VGNL+ + + A
Sbjct: 57 ITMALFLTIPLSFIAIFLFYTLFQ----RLRFKLPPGPRPWPVVGNLYDIKPVRFRCFAE 224
Query: 63 LSKTHGPIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFI 122
++++GPI+++ G +++S+S+ AK L+ +D L++R + +++D L +
Sbjct: 225 WAQSYGPIISVWFGSTLNVIVSNSKLAKEVLKENDQQLADRHRSRSAAKFSRDGQDLIWA 404
Query: 123 PVSHRWRDLRKICNNLLFSNKNLNANEQLRCDKIL----ELFNDIKKSMLKGEAVNVGRL 178
+ +RK+C LFS K + A +R D++ +FND S G+ + + +
Sbjct: 405 DYGPHYVKVRKVCTLELFSPKRIEALRPIREDEVTAMVESIFNDSTNSENLGKGILMRKY 584
Query: 179 AFKTTINLLSNTIYSVDLIRSA---DKAG-EFKEVIVSIMKEVGRPNLADCFPVLKVVDP 234
N ++ + + S D+ G EFK ++ + +K +A+ P L+ + P
Sbjct: 585 IGAVAFNNITRLAFGKRFVNSEGVMDEQGVEFKAIVANGLKLGASLAMAEHIPWLRWMFP 764
Query: 235 --------HGIRRRTGSYFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNG 286
HG RR ++ + ++EE + R +SG K +DA+L +
Sbjct: 765 LEEEAFAKHGARRD---------RLTRAIMEEHTQARHKSG-GAKQHFVDALLTLQEKY- 911
Query: 287 SEMYKDKIERLSVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEE 346
++ +D I L D+ AG DT +VEW MAEL++NP V KA+ EL+K IG +++ E
Sbjct: 912 -DLSEDTIIGLLWDMITAGMDTTAISVEWAMAELIKNPRVQQKAQEELDKVIGFERVMTE 1088
Query: 347 SHIERLPYLQAIVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPN 406
+ LPYLQ + KE RLHP PL+LP +A ++ + GY IPKGS V VNVWA RDP
Sbjct: 1089TDFSSLPYLQCVAKEALRLHPPTPLMLPHRANTNVKIGGYDIPKGSNVHVNVWAVARDPA 1268
Query: 407 LWDNPNLFSPERFLGSEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNW 466
+W + F PERFL ++D KG F L PFGAGRR+CPG L I ++ MLG L++ F W
Sbjct: 1269VWKDATEFRPERFLEEDVDMKGHDFRLLPFGAGRRVCPGAQLGINMVTSMLGHLLHHFCW 1448
Query: 467 ELEGGIKPEDMDMDETFGLTLEKTQPVLAV 496
G+ P ++DM E G+ P+ V
Sbjct: 1449APPEGVNPAEIDMAENPGMVTYMRTPLQVV 1538
>TC80035 weakly similar to GP|18252325|gb|AAL66194.1 cytochrome P450 {Pyrus
communis}, partial (49%)
Length = 1342
Score = 172 bits (437), Expect(2) = 1e-72
Identities = 93/242 (38%), Positives = 139/242 (57%), Gaps = 11/242 (4%)
Frame = +1
Query: 214 MKEVGRPNLADCFPVLKVVDPHGIRRRTGSYFERLMKIFKGLVEERLKQ-----RKESGY 268
M G NLAD P L + D G+ R ++ K F +VE+ +K +K+ +
Sbjct: 628 MNLAGSFNLADFLPWLSIFDMQGLTSR----MKKTSKAFDQIVEKIIKDHEQDMKKDPHH 795
Query: 269 NTKNDMLDAMLDNAQHNGSEMYKD-----KIERLSVDLFVAGTDTVTSTVEWTMAELLRN 323
D+L +++ + E K I+ + +D+ D+ ++T+EW M+ELLR+
Sbjct: 796 KDFIDILLSLMHKSMDPHDEEQKQVIDITNIKAIILDMIGGALDSASTTIEWAMSELLRH 975
Query: 324 PEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIVKEIFRLHPAVPLLLPRQAEVDLDM 383
P VM K ++ELE +G V+E+ E LPYL ++KE RL+PA PLL PR+ D +
Sbjct: 976 PNVMKKLQNELENVVGLNNKVKETDFENLPYLNMVIKETLRLYPAGPLLAPRECLEDATI 1155
Query: 384 HGYTIPKGSQVLVNVWANGRDPNLW-DNPNLFSPERFLGSEIDFKGRSFELTPFGAGRRI 442
GY I K S+V++N WA GRDP +W +N ++F PERF+ S ID +G F+L PFG+GRR
Sbjct: 1156DGYYIKKNSRVIINAWAIGRDPKIWTNNCDIFYPERFINSNIDLRGHDFQLIPFGSGRRR 1335
Query: 443 CP 444
CP
Sbjct: 1336CP 1341
Score = 119 bits (297), Expect(2) = 1e-72
Identities = 74/216 (34%), Positives = 117/216 (53%), Gaps = 1/216 (0%)
Frame = +3
Query: 4 ISCILLFLLTITFILFLANTIL-KPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPHQTLAT 62
+S + +L + FIL L++ + PN + PPGPK PI+GNL LG PH+TL +
Sbjct: 6 LSLAIPAILFLIFILILSSYLFFHPNQHQHHQKNPPGPKPFPIIGNLHILGKLPHRTLQS 185
Query: 63 LSKTHGPIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFI 122
LSK HGPIM+LKLG+I TI++SS E+A+ L+ HD S+R A + L
Sbjct: 186 LSKIHGPIMSLKLGKIPTIIVSSPESAEKFLKTHDAIFSSRPKLQASEHMFYGGKGLGLA 365
Query: 123 PVSHRWRDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKT 182
P WR+++K+C L S + +R +++ L + K+ + GE VN+ + +
Sbjct: 366 PYGSYWRNMKKLCTLHLLSGSKVEMFASMRSEELGMLIKSVDKAAVLGEVVNLSEIVGE- 542
Query: 183 TINLLSNTIYSVDLIRSADKAGEFKEVIVSIMKEVG 218
+++N Y + L + D + K VI S + E+G
Sbjct: 543 ---VIANITYKMVLGCNKDSDLDLK-VIYSRVYELG 638
>TC85609 weakly similar to SP|Q9LTL2|C72P_ARATH Cytochrome P450 71B25 (EC
1.14.-.-). [Mouse-ear cress] {Arabidopsis thaliana},
partial (33%)
Length = 1786
Score = 269 bits (687), Expect = 2e-72
Identities = 165/498 (33%), Positives = 261/498 (52%), Gaps = 9/498 (1%)
Frame = +1
Query: 11 LLTITFILFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKP-HQTLATLSKTHGP 69
L+ + F L L K KK LPPGPKGLP +GNL QL LSK +GP
Sbjct: 64 LILLPFALLLFFLFKKHKTSKKSTTLPPGPKGLPFIGNLHQLDSSVLGLNFYELSKKYGP 243
Query: 70 IMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFIPVSHRWR 129
I++LKLG T+V+SS++ AK ++ HD NR + + + F P WR
Sbjct: 244 IISLKLGSKQTVVVSSAKMAKEVMKTHDIEFCNRPALISHMKISYNGLDQIFAPYREYWR 423
Query: 130 DLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTINLLSN 189
+K+ S K ++ +R D++ + I ++ + +N+ L T L+
Sbjct: 424 HTKKLSFIHFLSVKRVSMFYSVRKDEVTRMIKKISENASSNKVMNMQDLLTCLTSTLVCK 603
Query: 190 TIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNL----ADCFPVLK-VVDP-HGIRRRTGS 243
T + R + G + + + KEV + AD P + +VD G R
Sbjct: 604 TAFG----RRYEGEGIERSMFQGLHKEVQDLLISFFYADYLPFVGGIVDKLTGKTSRLEK 771
Query: 244 YFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLD--NAQHNGSEMYKDKIERLSVDL 301
F+ ++++ +V+E L ++ ++D++DA+++ N + ++ + I+ L +++
Sbjct: 772 TFKVSDELYQSIVDEHLDPERKKLPPHEDDVIDALIELKNDPYCSMDLTAEHIKPLIMNM 951
Query: 302 FVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIVKE 361
A T+T+ + V W M L++NP M K + E+ K +EE +E+LPY +A++KE
Sbjct: 952 SFAVTETIAAAVVWAMTALMKNPRAMQKVQEEIRKVCAGKGFIEEEDVEKLPYFKAVIKE 1131
Query: 362 IFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSPERFLG 421
RL+P +P+LLPR+ + ++ GY IP + V VN A RDP +W +P F PERF+G
Sbjct: 1132SMRLYPILPILLPRETMTNCNIAGYDIPDKTLVYVNALAIHRDPEVWKDPEEFYPERFIG 1311
Query: 422 SEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPEDMDMDE 481
S+ID KG+ FEL PFG+GRRICPGL +AI + L+L L+ F+WE+ G K ED+D
Sbjct: 1312SDIDLKGQDFELIPFGSGRRICPGLNMAIATIDLVLSNLLYSFDWEMPEGAKREDIDTHG 1491
Query: 482 TFGLTLEKTQPVLAVPTK 499
GL K P+ V K
Sbjct: 1492QAGLIQHKKNPLCLVAKK 1545
>TC77891 weakly similar to SP|P93147|C81E_GLYEC Cytochrome P450 81E1 (EC
1.14.-.-) (Isoflavone 2'-hydroxylase) (P450 91A4) (CYP
GE-3). [Licorice], partial (91%)
Length = 1707
Score = 268 bits (684), Expect = 5e-72
Identities = 177/493 (35%), Positives = 255/493 (50%), Gaps = 10/493 (2%)
Frame = +1
Query: 7 ILLFLLTITFILFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPHQTLATLSKT 66
I LF L IT +F N +K LPPGP+ LPI+GNL QL H T TLS+
Sbjct: 55 ISLFFLIITLKVFF-------NTSRKFKNLPPGPQCLPIIGNLHQLKQPLHHTFHTLSQK 213
Query: 67 HGPIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRK--IPDAMKGYNQDRYSLPFIPV 124
+G I +L G +V+SS A+ +D L+NR + GYN + P
Sbjct: 214 YGQIFSLWFGSRLVVVVSSLTIAQECFTKNDIVLANRPHFLTGKYIGYNNTTVAQS--PY 387
Query: 125 SHRWRDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAV-----NVGRLA 179
WR+LR+I + + S+ LN+ ++R D+I+ L + + G +
Sbjct: 388 GDHWRNLRRILSIEILSSHRLNSFLEIRRDEIMRLIQKLAQKSYNGFTEVELRPMFSEMT 567
Query: 180 FKTTINLLSNT-IYSVDL-IRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHGI 237
F T + ++S Y D + ++A F+ +I ++ G N+ D L+ D G+
Sbjct: 568 FNTIMRMVSGKRYYGNDCDVSDVEEARLFRGIIKEVVSLGGANNVGDFLGFLRWFDFDGL 747
Query: 238 RRRTGSYFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNGSEMYKDKIER- 296
+R +R +GL++E G N M+D +L Q + E Y D+I +
Sbjct: 748 EKRLKKISKRTDAFLQGLIDEH-----RFGKRNSNTMIDHLLTQ-QQSQPEYYTDQIIKG 909
Query: 297 LSVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQ 356
L V + +AGTDT + T+EW M+ LL +PE+M KAK+EL+ IG + V+E I +LPYLQ
Sbjct: 910 LMVVMLLAGTDTSSVTIEWAMSNLLNHPEIMKKAKNELDTHIGHDRQVDEHDISKLPYLQ 1089
Query: 357 AIVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSP 416
+IV E RLH A PLL+P + D + GY IP+ + ++VN W RDPNLW +P F P
Sbjct: 1090SIVYETLRLHAAAPLLVPHLSSEDFSLGGYNIPQNTILMVNAWVIHRDPNLWSDPTCFKP 1269
Query: 417 ERFLGSEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPED 476
ERF + +G +L FG GRR CPG LA R L LGLLI CF W+ G E
Sbjct: 1270ERF-----EKEGEVNKLLSFGLGRRACPGENLAQRTEGLTLGLLIQCFEWKRIG---EEK 1425
Query: 477 MDMDETFGLTLEK 489
+DM E G+T+ K
Sbjct: 1426IDMVEAKGITVGK 1464
>TC87269 similar to PIR|T05940|T05940 cytochrome P450 83D1p - soybean
(fragment), partial (79%)
Length = 1878
Score = 267 bits (683), Expect = 6e-72
Identities = 162/509 (31%), Positives = 271/509 (52%), Gaps = 16/509 (3%)
Frame = +1
Query: 7 ILLFLLTITFILFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLG-HKPHQTLATLSK 65
+LL LL I ILF+ + I K + PPGPK LP++GNL QL PH +L LSK
Sbjct: 235 VLLSLLPI-LILFIIH-IYKIRSTSRASSTPPGPKPLPLIGNLHQLDPSSPHHSLWKLSK 408
Query: 66 THGPIMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFIPVS 125
+GPIM+L+LG I T+++SS++ A+ L+ HD ++R ++ + + L F P S
Sbjct: 409 HYGPIMSLQLGYIPTLIVSSAKMAEQVLKTHDLKFASRPSFLGLRKLSYNGLDLAFAPYS 588
Query: 126 HRWRDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTIN 185
WR++RK+C LFS++ +++ +R +++ +L + + + N+ + T
Sbjct: 589 PYWREMRKLCVQHLFSSQRVHSFRPVRENEVAQLIQKLSQYGGDEKGANLSEILMSLTNT 768
Query: 186 LLSNTIYSVDLI---------RSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDP-H 235
++ + + S K + ++ + +D FP+L +D
Sbjct: 769 IICKIAFGKTYVCDYEEEVELGSGQKRSRLQVLLNEAQALLAEFYFSDNFPLLGWIDRVK 948
Query: 236 GIRRRTGSYFERLMKIFKGLVEERLKQ--RKESGYNTKNDMLDAMLD--NAQHNGSEMYK 291
G + F+ L I++ ++++ + R ++ +D++D +L N ++
Sbjct: 949 GTLGKLDKTFKELDLIYQRVIDDHMDNSARPKTKEQEVDDIIDILLQMMNDHSLSFDLTL 1128
Query: 292 DKIERLSVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIER 351
D I+ + +++F+AGTDT ++ V W M L+ NP VM+K + E+ + E IE+
Sbjct: 1129DHIKAVLMNIFIAGTDTSSAIVVWAMTTLMNNPRVMNKVQMEIRNLYEDKDFINEDDIEK 1308
Query: 352 LPYLQAIVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNP 411
LPYL+A+VKE RL P PLL+PR+ + ++ GY I + V VN WA RDP W +P
Sbjct: 1309LPYLKAVVKETIRLFPPSPLLVPRETIENCNIDGYEIKPKTLVYVNAWAIARDPENWKDP 1488
Query: 412 NLFSPERFLGSEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGG 471
F PERF+ S +DFKG++FEL PFG+GRR+CP + + + + L L L+ F+W L G
Sbjct: 1489EEFYPERFIISSVDFKGKNFELIPFGSGRRMCPAMNMGVVTVELTLANLLQSFDWNLPHG 1668
Query: 472 I-KPEDMDMDETFGLTLEKTQPVLAVPTK 499
K + +D G+T+ K + VP K
Sbjct: 1669FDKEQVLDTQVKPGITMHKKIDLHLVPMK 1755
>TC87669 weakly similar to GP|3850630|emb|CAA10067.1 cytochrome P450 {Cicer
arietinum}, partial (86%)
Length = 1936
Score = 265 bits (676), Expect = 4e-71
Identities = 176/501 (35%), Positives = 261/501 (51%), Gaps = 15/501 (2%)
Frame = +3
Query: 11 LLTITFILFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKPHQTLATLSKTHGPI 70
LL+++FI+ + +LK ++ K LPPGP +PI+GNL L H H+T TLS+T+G I
Sbjct: 18 LLSLSFIITI-KILLKITSRRLK-NLPPGPPTIPIIGNLHHLKHPLHRTFTTLSQTYGDI 191
Query: 71 MTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKG----YNQDRYSLPFIPVSH 126
+L G +V+SS A +D L+NR P + G YN +L
Sbjct: 192 FSLWFGSRLVVVVSSPSLAHECFTKNDIILANR--PRFLTGKYIFYNYT--TLGSASYGD 359
Query: 127 RWRDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGE--------AVNVGRL 178
WR+LR+I + SN LN+ +R D+ L + K ++ + +
Sbjct: 360 HWRNLRRITTIDVLSNNRLNSFLGVRRDETNRLIQKLLKDVVSEGFGFTKVELRPRLTEM 539
Query: 179 AFKTTINLLSNTIYSVDL--IRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHG 236
F + ++S Y D + ++A +F+E+I +M +G N D P+L+VVD
Sbjct: 540 TFNAMMRMISGKRYYGDDGDVSDVEEAKQFREIISEMMSLLGANNKGDFLPLLRVVDLDN 719
Query: 237 IRRRTGSYFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNGSEMYKDK-IE 295
+ +R +R +GL+EE R+ + ++ M+D +L ++ E Y D I+
Sbjct: 720 LEKRCKRIAKRSNAFLEGLIEEH---RRGNIHSDGGTMIDHLLKLSESQ-PEYYSDHLIK 887
Query: 296 RLSVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYL 355
L + +AGTDT T+EW M+ELL +PEV+ KAK EL IGK KLV+E + +LPYL
Sbjct: 888 GLIQGMLLAGTDTSAVTIEWVMSELLNHPEVLKKAKEELGTQIGKNKLVDEQDLSKLPYL 1067
Query: 356 QAIVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFS 415
Q I+ E RLHP PLLLP + D + + +PK + +L NVW RDP W++ F
Sbjct: 1068QNIISETLRLHPPAPLLLPHYSSEDCTIGEFNVPKDTIILTNVWGIHRDPKHWNDALSFK 1247
Query: 416 PERFLGSEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPE 475
PERF E ++ FG GRR CPGL LA R + +GLLI CF WE E E
Sbjct: 1248PERFEKEE-----EVNKVMAFGLGRRACPGLSLAQRTVGFTVGLLIQCFEWERE---SEE 1403
Query: 476 DMDMDETFGLTLEKTQPVLAV 496
+DM E G+T+ P+ A+
Sbjct: 1404KLDMMEGKGITMPMKIPLRAM 1466
>TC84115 weakly similar to GP|17065916|emb|CAC80883. geraniol 10-hydroxylase
{Catharanthus roseus}, partial (33%)
Length = 785
Score = 263 bits (672), Expect(2) = 1e-70
Identities = 131/247 (53%), Positives = 178/247 (72%)
Frame = +1
Query: 129 RDLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTINLLS 188
RDLRKIC N LFS++ L+A+ LRC K+ EL +DI+KS L GEAV+VGR AFKT++N LS
Sbjct: 43 RDLRKICKNQLFSSQTLDASHALRCKKLQELLSDIEKSSLIGEAVDVGRAAFKTSLNFLS 222
Query: 189 NTIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNLADCFPVLKVVDPHGIRRRTGSYFERL 248
NT +S+D + SA + E+K ++ ++++ +G PNL D FPV K++DP GIR + +Y E+L
Sbjct: 223 NTFFSMDFVNSAGETDEYKGIVENLVRAIGTPNLVDFFPVFKMIDPQGIRGISATYVEKL 402
Query: 249 MKIFKGLVEERLKQRKESGYNTKNDMLDAMLDNAQHNGSEMYKDKIERLSVDLFVAGTDT 308
++I + +RLK R+E Y T NDMLD +L+ +Q NG +M KI+ L +DLFVAGTDT
Sbjct: 403 LQIIDSYIGKRLKLREEKDYVTNNDMLDNLLNISQENGQKMDNTKIKHLFLDLFVAGTDT 582
Query: 309 VTSTVEWTMAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIVKEIFRLHPA 368
+ T+E MAEL+ NP MSKAK EL++ IG G +EES I +LPYLQAI K+ RLH +
Sbjct: 583 TSYTIERAMAELIHNPHAMSKAKEELQEIIGIGNTIEESDIFKLPYLQAIXKKTLRLHXS 762
Query: 369 VPLLLPR 375
LLLPR
Sbjct: 763 AXLLLPR 783
Score = 21.9 bits (45), Expect(2) = 1e-70
Identities = 8/17 (47%), Positives = 11/17 (64%)
Frame = +3
Query: 117 YSLPFIPVSHRWRDLRK 133
+SLPF+PV W +K
Sbjct: 6 FSLPFMPVCDLWERPKK 56
>TC85610 similar to GP|21068674|emb|CAD31843. putative cytochrome P450
monooxygenase {Cicer arietinum}, complete
Length = 1578
Score = 172 bits (436), Expect(2) = 3e-70
Identities = 117/389 (30%), Positives = 195/389 (50%), Gaps = 10/389 (2%)
Frame = +2
Query: 11 LLTITFILFLANTILKPNHKKKKLPLPPGPKGLPIVGNLFQLGHKP-HQTLATLSKTHGP 69
L+ + F L L K KK LPPGPKGLP +GNL QL LSK +G
Sbjct: 17 LILLPFALLLFFLFKKHKTSKKSTTLPPGPKGLPFIGNLHQLDSSALGLNFYELSKKYGS 196
Query: 70 IMTLKLGQITTIVMSSSETAKSTLQIHDNFLSNRKIPDAMKGYNQDRYSLPFIPVSHRWR 129
++ LKLG TIV+SS++ AK ++ HD NR + ++ D F P WR
Sbjct: 197 LIYLKLGSRQTIVVSSAKMAKQVMKTHDIDFCNRPALISHMKFSYDGLDQFFSPYREYWR 376
Query: 130 DLRKICNNLLFSNKNLNANEQLRCDKILELFNDIKKSMLKGEAVNVGRLAFKTTINLLSN 189
+K+ S K + +R + ++ I + + VN+ L T ++
Sbjct: 377 HTKKLSFIHFLSVKRVVMFSSVRKYETTQMITKISEHASSNKVVNLHELLMCLTSAVVCR 556
Query: 190 TIYSVDLIRSADKAGEFKEVIVSIMKEVGRPNLA----DCFPVLK-VVDPH-GIRRRTGS 243
T + R D+A E + + ++KE ++ D P + +VD G+ R
Sbjct: 557 TAFGR---RFEDEAAE-RSMFHDLLKEAQEMTISFFYTDYLPFVGGIVDKFTGLMSRLEK 724
Query: 244 YFERLMKIFKGLVEERLKQRKESGYNTKNDMLDAMLD--NAQHNGSEMYKDKIERLSVDL 301
F+ L F+ + +E + ++ + D++DA+++ N + ++ + I+ L +++
Sbjct: 725 LFKILDGFFQSVFDEHIDPNRKKLPPHEEDVIDALIELKNDPYCSMDLSAEHIKPLIMNM 904
Query: 302 FVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTI-GKGKLVEESHIERLPYLQAIVK 360
+AGTDT+ + V W M L++NP VM K + E+ K GKG +EE +++LPY +A++K
Sbjct: 905 LLAGTDTIAAAVVWAMTALMKNPRVMQKVQEEIRKAYEGKG-FIEEEDVQKLPYFKAVIK 1081
Query: 361 EIFRLHPAVPLLLPRQAEVDLDMHGYTIP 389
E RL+P++P+LLPR+ D+ GY IP
Sbjct: 1082ESMRLYPSLPVLLPRETMKKCDIEGYEIP 1168
Score = 111 bits (278), Expect(2) = 3e-70
Identities = 51/106 (48%), Positives = 70/106 (65%)
Frame = +3
Query: 394 VLVNVWANGRDPNLWDNPNLFSPERFLGSEIDFKGRSFELTPFGAGRRICPGLPLAIRLL 453
V +N WA RDP W +P F PERF+GS+ID KG+ FEL PFG+GRR+CPGL +AI +
Sbjct: 1182 VYINAWAIHRDPEAWKDPEEFYPERFIGSDIDLKGQDFELIPFGSGRRVCPGLNMAIATV 1361
Query: 454 FLMLGLLINCFNWELEGGIKPEDMDMDETFGLTLEKTQPVLAVPTK 499
L+L L+ F+WE+ G+K E++D+D GL K P+ + K
Sbjct: 1362 DLVLANLLYLFDWEMPEGVKWENIDIDGLPGLVQHKKNPLCLIAKK 1499
>CA920634 weakly similar to SP|O64636|C7C1 Cytochrome P450 76C1 (EC
1.14.-.-). [Mouse-ear cress] {Arabidopsis thaliana},
partial (30%)
Length = 720
Score = 257 bits (657), Expect = 6e-69
Identities = 123/183 (67%), Positives = 147/183 (80%)
Frame = -3
Query: 317 MAELLRNPEVMSKAKSELEKTIGKGKLVEESHIERLPYLQAIVKEIFRLHPAVPLLLPRQ 376
M EL+RNPE M KAK EL K IG +EES I +LPYLQAI+KE RLHP VPLLLPR+
Sbjct: 715 MTELIRNPEAMLKAKKELGKMIGCSVPLEESDISKLPYLQAIIKETLRLHPPVPLLLPRK 536
Query: 377 AEVDLDMHGYTIPKGSQVLVNVWANGRDPNLWDNPNLFSPERFLGSEIDFKGRSFELTPF 436
AE ++D+ GYTIPK +QV+VNVW GRDP +W NP LFSPERFLGS+ID KGR+FEL PF
Sbjct: 535 AERNVDIGGYTIPKDAQVMVNVWTIGRDPTIWLNPTLFSPERFLGSDIDVKGRNFELVPF 356
Query: 437 GAGRRICPGLPLAIRLLFLMLGLLINCFNWELEGGIKPEDMDMDETFGLTLEKTQPVLAV 496
GAGRRICPGL LA R+L LMLG L+N F+W+LEG +KPEDMDM++ FG+TL+K QP+ V
Sbjct: 355 GAGRRICPGLQLANRMLLLMLGSLVNSFDWKLEGDMKPEDMDMNDKFGITLQKAQPLRVV 176
Query: 497 PTK 499
P +
Sbjct: 175 PVR 167
>TC83545 weakly similar to GP|7406712|emb|CAB85635.1 putative
ripening-related P-450 enzyme {Vitis vinifera}, partial
(36%)
Length = 1243
Score = 256 bits (653), Expect = 2e-68
Identities = 118/207 (57%), Positives = 156/207 (75%)
Frame = +2
Query: 281 NAQHNGSEMYKDKIERLSVDLFVAGTDTVTSTVEWTMAELLRNPEVMSKAKSELEKTIGK 340
N + +++ ++++ L DLF AGTDT +ST+EW MAELL NPE ++KA+ EL K IGK
Sbjct: 29 NIEETTNKLSRNEMVHLFQDLFTAGTDTTSSTIEWVMAELLGNPEKLAKARKELCKEIGK 208
Query: 341 GKLVEESHIERLPYLQAIVKEIFRLHPAVPLLLPRQAEVDLDMHGYTIPKGSQVLVNVWA 400
+ +EESHI LP+LQA+VKE FRLHPA PLLLP + + +L++ G+ +PK +QVLVNVWA
Sbjct: 209 DETIEESHISMLPFLQAVVKETFRLHPAAPLLLPHKCDENLNISGFNVPKNAQVLVNVWA 388
Query: 401 NGRDPNLWDNPNLFSPERFLGSEIDFKGRSFELTPFGAGRRICPGLPLAIRLLFLMLGLL 460
GRDP +W+NPN F PERFL +I++KG +FEL PFGAG+RICPGLPLA R + L++ L
Sbjct: 389 MGRDPTIWENPNKFEPERFLERDINYKGNNFELIPFGAGKRICPGLPLAHRSVHLIVASL 568
Query: 461 INCFNWELEGGIKPEDMDMDETFGLTL 487
+ F W L G+ PEDM MDE FG+TL
Sbjct: 569 LRNFEWTLADGLNPEDMSMDERFGVTL 649
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,694,803
Number of Sequences: 36976
Number of extensions: 205837
Number of successful extensions: 1664
Number of sequences better than 10.0: 261
Number of HSP's better than 10.0 without gapping: 1484
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1506
length of query: 502
length of database: 9,014,727
effective HSP length: 100
effective length of query: 402
effective length of database: 5,317,127
effective search space: 2137485054
effective search space used: 2137485054
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0237.2


