

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
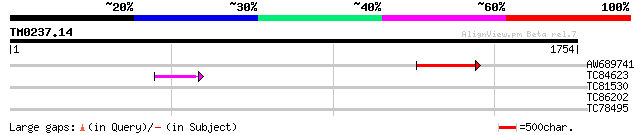
Query= TM0237.14
(1754 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW689741 similar to GP|9759369|dbj gene_id:MYJ24.10~ref|NP_05517... 317 2e-86
TC84623 similar to GP|9759369|dbj|BAB09828.1 gene_id:MYJ24.10~re... 82 2e-15
TC81530 similar to GP|10177440|dbj|BAB10736. gb|AAC55944.1~gene_... 33 0.97
TC86202 similar to GP|3738234|dbj|BAA33796.1 sulfite reductase {... 32 2.8
TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 ... 31 4.8
>AW689741 similar to GP|9759369|dbj gene_id:MYJ24.10~ref|NP_055178.1~similar to
unknown protein {Arabidopsis thaliana}, partial (1%)
Length = 651
Score = 317 bits (813), Expect = 2e-86
Identities = 154/203 (75%), Positives = 174/203 (84%), Gaps = 3/203 (1%)
Frame = +2
Query: 1257 SQGGPSSEWIKIFWKSFRGSQEELSLFSDWPLIPAFLGRPVLCRVRERHLVFIPPPLVQR 1316
SQGGPSSEW++IFWK+F+GSQEELSLFSDWPLIPAFLGRPVLCRVRERHLVFIPPPL
Sbjct: 11 SQGGPSSEWVRIFWKNFKGSQEELSLFSDWPLIPAFLGRPVLCRVRERHLVFIPPPLEHP 190
Query: 1317 TLQSGILDRESTENHVGGVSVSRDDTS---VAEAYTSAFDRLKISYPWLLSLLNQCNVPI 1373
T S IL+ ESTE++ GV +S D++S +AE+Y+SAF+RLKISYPWLL +LNQCN+PI
Sbjct: 191 TSTSQILETESTESYADGVRISSDNSSEAELAESYSSAFERLKISYPWLLPMLNQCNIPI 370
Query: 1374 FDEAFIDCATSSNCFSMPGGSLGQVIASKLVAAKQAGYFTEPNNLSASSCDALFSLFCDE 1433
FDEAFIDCA SSNCFS P SLGQVIASKLVA KQAGYFTEP + S S+CDALFSLF E
Sbjct: 371 FDEAFIDCAASSNCFSTPERSLGQVIASKLVAVKQAGYFTEPTDFSTSNCDALFSLFSGE 550
Query: 1434 LFSNGFHYAQEEIEILRSLPIYK 1456
+SN YAQEEIE+LRSLPIYK
Sbjct: 551 FYSNSVRYAQEEIEVLRSLPIYK 619
>TC84623 similar to GP|9759369|dbj|BAB09828.1
gene_id:MYJ24.10~ref|NP_055178.1~similar to unknown
protein {Arabidopsis thaliana}, partial (3%)
Length = 634
Score = 81.6 bits (200), Expect = 2e-15
Identities = 52/155 (33%), Positives = 79/155 (50%), Gaps = 3/155 (1%)
Frame = +3
Query: 449 KVSELLALYGNSEFLLFDLLELADCCKAKKLHLIYDKREHPRQSLLQHNLGEFQGPALVA 508
++ E+L Y +L +L++ AD A + L D+R H R+SLL +L ++QGPAL+A
Sbjct: 168 RIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHGRESLLSDSLSQWQGPALLA 347
Query: 509 IFEGACLSREEFSNFQLLPPWKLRG---NTLNYGLGLVGCYSICDLLSVVSGGYFYMFDP 565
+ A S E+F + + G T +G+G Y + DL S VSG Y +FDP
Sbjct: 348 -YNDAVFSEEDFVSISKIGGSSKHGQASKTGRFGVGFNSVYHLTDLPSFVSGKYLVLFDP 524
Query: 566 RGLVLSAPSSNAPSGKMFSLIGTDLAQRFGDQFSP 600
+G+ L S+ P GK + + DQF P
Sbjct: 525 QGVYLPRVSAXNP-GKRIDFTSSSALSFYXDQFFP 626
>TC81530 similar to GP|10177440|dbj|BAB10736.
gb|AAC55944.1~gene_id:K9H21.2~similar to unknown protein
{Arabidopsis thaliana}, partial (24%)
Length = 1135
Score = 33.1 bits (74), Expect = 0.97
Identities = 17/42 (40%), Positives = 24/42 (56%)
Frame = -1
Query: 911 PSSSTAVLKADWECLKERVIHPFYSRIVDLPVWQLYSGNLVK 952
PS+S+ VL A + V HPF ++L VW +G+LVK
Sbjct: 475 PSASSQVLLAQS*AFQGHVFHPFQGLSLELSVWCQQTGSLVK 350
>TC86202 similar to GP|3738234|dbj|BAA33796.1 sulfite reductase {Nicotiana
tabacum}, partial (91%)
Length = 2454
Score = 31.6 bits (70), Expect = 2.8
Identities = 17/58 (29%), Positives = 25/58 (42%)
Frame = +2
Query: 209 DCPNIGVLSRQLIELSKSYQQLKTHSLLDPDFDVKLQKEIPCLYSKLQEYINTDDFND 266
D PN+ + QLI+ SYQQ + ++ + PC Q Y+ DD D
Sbjct: 386 DAPNLSEAATQLIKFHGSYQQYNRDERGSRTYSFMIRTKNPCGKVSNQLYLTMDDLAD 559
>TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 - alfalfa,
partial (97%)
Length = 1049
Score = 30.8 bits (68), Expect = 4.8
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Frame = -2
Query: 623 SDCLKVGHDVASNRIK-HITDVFMEHGSRTLLFLKSVLQVSISTWEEGHSHPCQNFSISI 681
++CLK ++V SN++ HI + + G L + +VSI ++ +HP N +I
Sbjct: 961 NECLKNANNVCSNKLPYHIEKIIIYRGKNNNLSSEEKTEVSI*NSKDRKAHPNTNPTILC 782
Query: 682 DPSSS 686
D + S
Sbjct: 781 DHNFS 767
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 59,258,511
Number of Sequences: 36976
Number of extensions: 910919
Number of successful extensions: 4151
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4078
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4148
length of query: 1754
length of database: 9,014,727
effective HSP length: 110
effective length of query: 1644
effective length of database: 4,947,367
effective search space: 8133471348
effective search space used: 8133471348
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0237.14


