

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
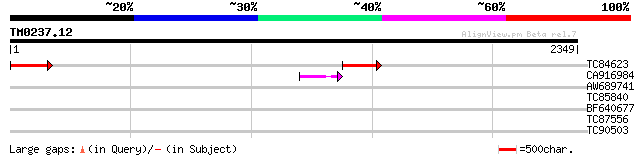
Query= TM0237.12
(2349 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84623 similar to GP|9759369|dbj|BAB09828.1 gene_id:MYJ24.10~re... 326 6e-89
CA916984 similar to GP|9759369|db gene_id:MYJ24.10~ref|NP_055178... 44 6e-04
AW689741 similar to GP|9759369|dbj gene_id:MYJ24.10~ref|NP_05517... 41 0.005
TC85840 similar to SP|P35016|ENPL_CATRO Endoplasmin homolog prec... 35 0.34
BF640677 weakly similar to GP|8843797|dbj contains similarity to... 32 2.2
TC87556 similar to GP|14517530|gb|AAK62655.1 At1g48370/F11A17_27... 32 2.2
TC90503 weakly similar to GP|22655264|gb|AAM98222.1 unknown prot... 30 8.5
>TC84623 similar to GP|9759369|dbj|BAB09828.1
gene_id:MYJ24.10~ref|NP_055178.1~similar to unknown
protein {Arabidopsis thaliana}, partial (3%)
Length = 634
Score = 326 bits (836), Expect = 6e-89
Identities = 160/175 (91%), Positives = 167/175 (95%)
Frame = +3
Query: 3 SPSPESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSH 62
SP+PESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSH
Sbjct: 108 SPAPESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSH 287
Query: 63 AGDSLLSSSLSQWQGPALLAYNDAVFTEDDFISISKIGGSSKHGQASKTGRFGVGFNSVY 122
+SLLS SLSQWQGPALLAYNDAVF+E+DF+SISKIGGSSKHGQASKTGRFGVGFNSVY
Sbjct: 288 GRESLLSDSLSQWQGPALLAYNDAVFSEEDFVSISKIGGSSKHGQASKTGRFGVGFNSVY 467
Query: 123 HLTDLPSFVSGKYVVLFDPQGVYLPRVSAANPGKRIDFTGSSALSLYKDQFSPYC 177
HLTDLPSFVSGKY+VLFDPQGVYLPRVSA NPGKRIDFT SSALS Y DQF P+C
Sbjct: 468 HLTDLPSFVSGKYLVLFDPQGVYLPRVSAXNPGKRIDFTSSSALSFYXDQFFPFC 632
Score = 158 bits (400), Expect = 2e-38
Identities = 78/162 (48%), Positives = 105/162 (64%)
Frame = +3
Query: 1380 EAFGQHEALTTRLKHILEMYADGPGTLFELVQNAEDAGASEVIFLLDKSQYGTSSVLSPE 1439
E FGQ LT R++ +L Y +G L EL+QNA+DAGA+ V LD+ +G S+LS
Sbjct: 135 EDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHGRESLLSDS 314
Query: 1440 MADWQGPALYCFNDSVFTPQDLYAISRIGQESKLEKAFAIGRFGLGFNCVYHFTDIPMFV 1499
++ WQGPAL +ND+VF+ +D +IS+IG SK +A GRFG+GFN VYH TD+P FV
Sbjct: 315 LSQWQGPALLAYNDAVFSEEDFVSISKIGGSSKHGQASKTGRFGVGFNSVYHLTDLPSFV 494
Query: 1500 SGENIVMFDPHASNLPGISPSHPGLRIKFAGRKILEQFPDQF 1541
SG+ +V+FDP LP +S +PG RI F L + DQF
Sbjct: 495 SGKYLVLFDPQGVYLPRVSAXNPGKRIDFTSSSALSFYXDQF 620
>CA916984 similar to GP|9759369|db gene_id:MYJ24.10~ref|NP_055178.1~similar to
unknown protein {Arabidopsis thaliana}, partial (3%)
Length = 696
Score = 44.3 bits (103), Expect = 6e-04
Identities = 42/187 (22%), Positives = 80/187 (42%), Gaps = 7/187 (3%)
Frame = +1
Query: 1199 GDGFATSDEVVLDGPLHLAPYIRVIPVDLAVFKNLFLELGIREFLQPSDYVNILFRMANK 1258
G T+D + ++L+P+ +P F + +LG+++ L S ++L +
Sbjct: 37 GTRLVTADALFARLMINLSPFAFELPAVYLPFAKILKDLGLQDVLTLSAAKDLLLNLQKA 216
Query: 1259 KGSSPLDTQEIRAVMLIVHHLAEVYLHGQ-------KVQLYLPDVSGRLFLAGDLVYNDA 1311
G L+ E+RAVM I++ + + G K ++ +PD RL + VY D+
Sbjct: 217 CGYQHLNPNELRAVMEILNFICDQIGEGNTFGRYDWKSEVIVPDDGCRLVHSTSCVYVDS 396
Query: 1312 PWLLGSEDPDGSFGNAPSVTWNAKRTVQKFVHGNISNDVAEKLGVRSLRRMLLAESADSM 1371
DGS + +FVH ++ V LG++ L +++ E ++
Sbjct: 397 ---------DGS-----RYVKCIDTSRIRFVHADLPERVCIVLGIKKLSDVVIEELDENH 534
Query: 1372 NFGLSGA 1378
N G+
Sbjct: 535 NLETLGS 555
>AW689741 similar to GP|9759369|dbj gene_id:MYJ24.10~ref|NP_055178.1~similar to
unknown protein {Arabidopsis thaliana}, partial (1%)
Length = 651
Score = 41.2 bits (95), Expect = 0.005
Identities = 14/35 (40%), Positives = 23/35 (65%)
Frame = +2
Query: 2083 GIHGQPSLEWLQLLWNYLKANCDDLLMFSKWPILP 2117
G G PS EW+++ W K + ++L +FS WP++P
Sbjct: 8 GSQGGPSSEWVRIFWKNFKGSQEELSLFSDWPLIP 112
Score = 37.7 bits (86), Expect = 0.053
Identities = 14/34 (41%), Positives = 20/34 (58%)
Frame = +2
Query: 675 PTSSWFVLFWQYLGKQSEILPLFKDWPILPSTSG 708
P+S W +FW+ E L LF DWP++P+ G
Sbjct: 23 PSSEWVRIFWKNFKGSQEELSLFSDWPLIPAFLG 124
>TC85840 similar to SP|P35016|ENPL_CATRO Endoplasmin homolog precursor
(GRP94 homolog). [Rosy periwinkle Madagascar
periwinkle], partial (92%)
Length = 2791
Score = 35.0 bits (79), Expect = 0.34
Identities = 35/127 (27%), Positives = 58/127 (45%), Gaps = 17/127 (13%)
Frame = +1
Query: 23 RIREVLLN--YPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGD---SLLSSSLSQWQG 77
R+ ++++N Y L+ELI NA DA L L + G+ + L + +
Sbjct: 307 RLMDIIINSLYSNKDIFLRELISNASDALDKIRFLSLTDKDILGEGDNAKLEIQIKLDKE 486
Query: 78 PALLAYNDAVF--TEDDFI----SISKIGGSS------KHGQASKTGRFGVGFNSVYHLT 125
+L+ D T++D + +I+K G S+ G + G+FGVGF SVY +
Sbjct: 487 KKILSIRDRGIGMTKEDLVKNLGTIAKSGTSAFVEKMQTSGDLNLIGQFGVGFYSVYLVA 666
Query: 126 DLPSFVS 132
D +S
Sbjct: 667 DYVEVIS 687
>BF640677 weakly similar to GP|8843797|dbj contains similarity to unknown
protein~gb|AAF22257.1~gene_id:MZN1.21 {Arabidopsis
thaliana}, partial (40%)
Length = 674
Score = 32.3 bits (72), Expect = 2.2
Identities = 18/62 (29%), Positives = 30/62 (48%), Gaps = 2/62 (3%)
Frame = +3
Query: 2054 FLSCQLLEKLLVKLLPVEWQHASLVSWTPGIHGQPSLEWLQLL--WNYLKANCDDLLMFS 2111
F+ L KL++ +L +ASL+SW I L W ++ W++ + NC + S
Sbjct: 120 FIGGSLRYKLILDILQSLLPNASLLSWLFRIGWMGKLLWTEIFQKWSHCRRNCKRI*CLS 299
Query: 2112 KW 2113
W
Sbjct: 300 MW 305
>TC87556 similar to GP|14517530|gb|AAK62655.1 At1g48370/F11A17_27 {Arabidopsis
thaliana}, partial (48%)
Length = 1309
Score = 32.3 bits (72), Expect = 2.2
Identities = 23/89 (25%), Positives = 43/89 (47%)
Frame = +1
Query: 1709 KGTSEASTSNSHNFVPWACVAAYLNSVKHGEDLVDSAEVEDDCLVSSDLFQFASLPMHPR 1768
K + T S P+A V Y N G S + D+CL+ +F A++ ++
Sbjct: 658 KAFPDLGTHKSQYPAPYAIV--YRNMAILGVQGFSS--LPDNCLLLCYIFFGAAIVVNLI 825
Query: 1769 ENFEGRAFCFLPLPISTGLPAHVNAYFEL 1797
++ G+ F+PLP++ +P ++ YF +
Sbjct: 826 KDLVGKVGRFIPLPMAMAIPFYLGPYFAI 912
>TC90503 weakly similar to GP|22655264|gb|AAM98222.1 unknown protein
{Arabidopsis thaliana}, partial (8%)
Length = 846
Score = 30.4 bits (67), Expect = 8.5
Identities = 20/54 (37%), Positives = 28/54 (51%)
Frame = +2
Query: 808 MDEFNIRFCRRLPIYQVYHREPTQDSQFSDLENPRKYLPPLDVPEFILVGIEFI 861
+D N R C P YQVYH +F+ L+ +K L +V E I V +EF+
Sbjct: 539 LDSVN-RSCSFHPSYQVYH----HSIEFAGLKVTKKLLEKKNV*EIINVALEFV 685
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 76,427,698
Number of Sequences: 36976
Number of extensions: 1166940
Number of successful extensions: 5317
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 5125
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5315
length of query: 2349
length of database: 9,014,727
effective HSP length: 112
effective length of query: 2237
effective length of database: 4,873,415
effective search space: 10901829355
effective search space used: 10901829355
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0237.12


