

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0228a.4
(1904 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
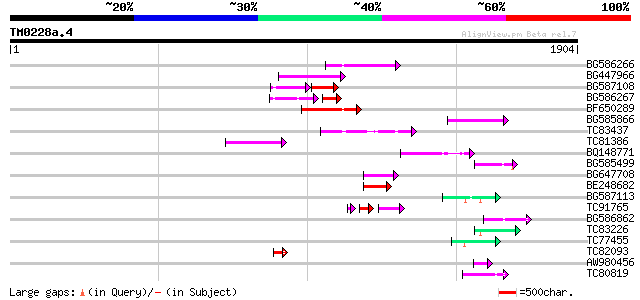
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 166 1e-40
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 154 2e-37
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 89 4e-35
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 112 9e-34
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 136 7e-32
BG585866 128 2e-29
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 105 2e-22
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 92 2e-18
BQ148771 91 4e-18
BG585499 79 2e-14
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 78 3e-14
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 73 9e-13
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 68 3e-11
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 42 2e-09
BG586862 55 3e-07
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 51 5e-06
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 50 1e-05
TC82093 48 3e-05
AW980456 48 3e-05
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 44 6e-04
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 166 bits (419), Expect = 1e-40
Identities = 93/255 (36%), Positives = 143/255 (55%), Gaps = 3/255 (1%)
Frame = -3
Query: 1059 KLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHT 1118
++ + R I+ CN YK+I K+L R +PLL ++SP Q +F+ GR DN+++ +++H
Sbjct: 781 RVSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILH- 605
Query: 1119 IHTRKTGGGL---VALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSV 1175
+ R++G +A+K D+ KAYDR+ W+FLR L GF + IM V T + S
Sbjct: 604 -YLRQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSF 428
Query: 1176 LWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGIS 1235
L NG RGLRQGDPLSPYLF+LC E L+ Q + +G V+V+++ I+
Sbjct: 427 LINGGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPIN 248
Query: 1236 HLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSG 1295
HL FADD + F +++ ++ + + ASG +N KS + FS Q + G
Sbjct: 247 HLLFADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKG 68
Query: 1296 LVGISRASNLGKYLG 1310
+ I++ GKYLG
Sbjct: 67 ELKIAKEGGTGKYLG 23
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 154 bits (390), Expect = 2e-37
Identities = 86/223 (38%), Positives = 117/223 (51%)
Frame = +2
Query: 903 WVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTNLFSLDVQV 962
W++ GD+NTKFFH++ RRK N+I L E GNWC ++ ++ NLF+
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPT 184
Query: 963 RRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHIL 1022
E + E + EEV AI M KAPGPDG A F+++YWHI+
Sbjct: 185 AIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIV 364
Query: 1023 GEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVN 1082
G+++ MV N EL T +VLIPK NP KD RPISLCNV K+ITKV+ N
Sbjct: 365 GKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIAN 544
Query: 1083 RFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTG 1125
R + L ++ Q +F+ GR DN ++A V R+ G
Sbjct: 545 RVKQTLPDVIDVEQSAFVQGRLITDNALIAWSVSIG*RXRRKG 673
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 88.6 bits (218), Expect(2) = 4e-35
Identities = 51/134 (38%), Positives = 72/134 (53%)
Frame = +2
Query: 876 LERDLQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDG 935
L+ LQ+ Y D EE W QKSR W GD N KF+HA T R RN+I GL+ DG
Sbjct: 8 LKEKLQEAYKD----EEDYWQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDG 175
Query: 936 NWCTDPSTLQEEAKRFYTNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSA 995
NW T+ +++ A ++ +LF + ++ T +I + +L+ + EEVR A
Sbjct: 176 NWITEEQGVEKVAVDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLATEEEVRLA 355
Query: 996 IMSMKSFKAPGPDG 1009
+ M KAPGPDG
Sbjct: 356 LFIMHPEKAPGPDG 397
Score = 80.1 bits (196), Expect(2) = 4e-35
Identities = 40/89 (44%), Positives = 55/89 (60%)
Frame = +3
Query: 1014 FYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSY 1073
F++ WHI+ DL MV S +G + L T + LIPK PT++ +LRPISLCNV Y
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 1074 KLITKVLVNRFRPLLHKLVSPLQGSFILG 1102
K+I+KVL R + L L+S Q +F+ G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 112 bits (279), Expect(2) = 9e-34
Identities = 65/169 (38%), Positives = 92/169 (53%), Gaps = 2/169 (1%)
Frame = +3
Query: 871 ESLARLERDLQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGL 930
E L+ L +L +EY + EE+ W QKSR NW+R GDRNTKFFHA T RR +N+I L
Sbjct: 75 EELSLLRSELNEEY----HNEEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSL 242
Query: 931 FLEDG-NWCTDPSTLQEEAKRFYTNLFSLDVQVRRNRIMENQTPTIPMEERD-KLVAPVS 988
+D W + + F S DV + N P I EE++ +L+A +S
Sbjct: 243 IDDDDKEWFVEEDLGRLADSHFKLLYSSEDVGITLED--WNSIPAIVTEEQNAQLMAQIS 416
Query: 989 LEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNG 1037
EEVR A+ + K PGPDG + +F++Q+W +G+DL M + G
Sbjct: 417 REEVREAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTG 563
Score = 52.0 bits (123), Expect(2) = 9e-34
Identities = 25/63 (39%), Positives = 41/63 (64%)
Frame = +1
Query: 1050 LIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNI 1109
L+PK +L + RPISLCNV+YK+++KVL R + +L +++ Q +F + DNI
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 1110 ILA 1112
++A
Sbjct: 781 LIA 789
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 136 bits (343), Expect = 7e-32
Identities = 72/200 (36%), Positives = 122/200 (61%)
Frame = +3
Query: 980 RDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGA 1039
+D L + + EV++A+ SM S KAPG DG++ +F+K W+I+G+ + + F+ G
Sbjct: 3 QDLLCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFM 182
Query: 1040 NPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSF 1099
+ T + L+PK N T +K+ RPI+ C+V YK+I+K+L +R + +L+ +VS Q +F
Sbjct: 183 PKIINCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAF 362
Query: 1100 ILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPR 1159
+ GR DNIIL+ E++ + ++RK +KIDL KAYD W F++ + + GFP +
Sbjct: 363 VKGRVIFDNIILSHELVKS-YSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYK 539
Query: 1160 TVDLIMWGVRTPAVSVLWNG 1179
V+ +M + T + + NG
Sbjct: 540 FVNWVMAXLTTASYTFNXNG 599
>BG585866
Length = 828
Score = 128 bits (321), Expect = 2e-29
Identities = 67/209 (32%), Positives = 110/209 (52%), Gaps = 3/209 (1%)
Frame = +3
Query: 1469 ILKARDSVSDGFGPRLGAGSSSLWYSNWLGTGKIASRIPFVHITDTTLTVADIWRQGAWN 1528
I++ ++ + G+ R G+G+SS WY+NW G + ++ PFV I D LTV D++ G +
Sbjct: 84 IIRDKNVLKSGYTWRAGSGNSSFWYTNWSSLGLLGTQAPFVDIHDLHLTVKDVFTTGGQH 263
Query: 1529 FDRLYTLLPQEIREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQFL---NEDGLPE 1585
LYT+LP +I E I++ + + D +W S+ +Y+A S Y ++ E
Sbjct: 264 TQSLYTILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWILSQTETVNYN 443
Query: 1586 TGFWKLIWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLR 1645
W IW+ PEK +FFLWL +A+PT + + ++ +A C RCG E H +R
Sbjct: 444 NSSWSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHEESFFHCVR 623
Query: 1646 RCPGSVWVRGQLGRLLPWISDSATFRHWL 1674
C S + ++G P S++ + WL
Sbjct: 624 DCRFSKIIWHKIGFSSPDFFSSSSVQDWL 710
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 105 bits (262), Expect = 2e-22
Identities = 97/336 (28%), Positives = 147/336 (42%), Gaps = 15/336 (4%)
Frame = +2
Query: 1045 ATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRG 1104
+T + LIPKVDNP +L D RPISL YK++ K+L NR R ++ ++S Q +F+ R
Sbjct: 56 STFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKNRQ 235
Query: 1105 TRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFG-----FPPR 1159
+ + L Q + + L+ + K + + W R G F
Sbjct: 236 ILEMVFL*QMRLW-----------MRLR-N*RKIFCCLRWILKRLITLSIGLIWILF*VG 379
Query: 1160 TVDLIMWG------VRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQI 1213
L++W V T SVL NGS P+ L + L Q + Y F
Sbjct: 380 MSFLVLWRKWIKECVSTATTSVLVNGS--PTNVLMKSLVQTQLFTRYSF----------- 520
Query: 1214 QNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVN 1273
G PV VS HL FA+D LL + +R ++ L F SG+KVN
Sbjct: 521 ------GVVNPVVVS-------HLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVN 661
Query: 1274 LEKSRMLFSKVVGQRRQRELSGLVGISRASNLGK----YLGLPLLQGRVKRDHFATIIEK 1329
KS ++ + S L + +GK YLG+P+ + + I+ +
Sbjct: 662 FHKSGLVCVNIAPSWLSEAASVL-----SWKVGKVPFLYLGMPIEGNSRRLSFWEPIVNR 826
Query: 1330 VNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQS 1365
+ RL W + L+ GR+ L KS+L+SL V+A+ S
Sbjct: 827 IKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYALPS 934
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 92.0 bits (227), Expect = 2e-18
Identities = 63/210 (30%), Positives = 92/210 (43%), Gaps = 6/210 (2%)
Frame = -1
Query: 724 NFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSDHSPLL 783
N L GS FTW GR KRLDR++ + L ++ SDH P+L
Sbjct: 630 NLFHLPTRGSAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPIL 451
Query: 784 LRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGN-----LQDKLQRVQEAATE 838
+ + F+F W +HP +++ W Q L KL+ ++E
Sbjct: 450 FELQTQN-IQFSSSFKFMKMWSAHPDCINIIKQCWANQVVGCPMFVLNQKLKNLKEVLKV 274
Query: 839 FNKTVFGNIHRRKRRVERRLQGIQQEHDWRD-SESLARLERDLQKEYGDILYQEEMLWFQ 897
+NK FGN+H + + L IQ + D S+ L E+ Q L EE+ W +
Sbjct: 273 WNKNTFGNVHSQVDNAYKELDDIQVKIDSIGYSDVLMDQEKAAQLNLESALNIEEVFWHE 94
Query: 898 KSRENWVRFGDRNTKFFHAQTVVRRKRNKI 927
KS+ NW GDRNT FFH ++R + I
Sbjct: 93 KSKVNWHCEGDRNTAFFHRVAKIKRTSSLI 4
>BQ148771
Length = 680
Score = 90.9 bits (224), Expect = 4e-18
Identities = 63/252 (25%), Positives = 116/252 (46%), Gaps = 3/252 (1%)
Frame = -3
Query: 1313 LLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSV 1372
L++ K+ F+ + +V+ LA+WK N L+ A RV LAKS++ ++P++ M + +P +
Sbjct: 633 LVRKSTKKSRFS-VYYQVHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKAC 457
Query: 1373 CNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFL 1432
+ K+ R +WG +R +H V W+ +++ K LGLR N+A + KL S
Sbjct: 456 IEEIQKLQRKFVWGDTEVSRRYHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIY 277
Query: 1433 NEKGKLWVQALSQKYIKSGSILTEEL---RPGGSYVWRGILKARDSVSDGFGPRLGAGSS 1489
+ L + + KY +S S+ EE+ +P S +W+ ++K
Sbjct: 276 SGSNSLCTEVMRGKYQRSESL--EEIFLEKPTDSSLWKALVK------------------ 157
Query: 1490 SLWYSNWLGTGKIASRIPFVHITDTTLTVADIWRQGAWNFDRLYTLLPQEIREEISSVQI 1549
LW + + D G WN+++L LP ++ + I ++
Sbjct: 156 -LW-------------------PEIERNLVD--SNGNWNWEKLKQWLPFDVLQRIMAIAT 43
Query: 1550 PSIQTGEDRILW 1561
P G D+++W
Sbjct: 42 PHENLGSDKLIW 7
>BG585499
Length = 792
Score = 78.6 bits (192), Expect = 2e-14
Identities = 51/158 (32%), Positives = 73/158 (45%), Gaps = 15/158 (9%)
Frame = +3
Query: 1562 REASDDIYSASSAYQFLNEDGLPETGFWKLIWKASAPEKVRFFLWLVGRDALPTNAKRHR 1621
R S + S+Y L D WK++W P + + F+WLV + TN +R R
Sbjct: 144 RGTSTHQFKIKSSYNLLV*DQSIVDCDWKMLWGWRGPHRTQTFMWLVAHGCILTNYRRSR 323
Query: 1622 HHLATTASCIRCGAVVEDAEHVLRRC-PGS-VWVRGQLGRLLP--WISDSATF---RHWL 1674
A+C CG E HVL C P S VW+ RL+P WI++ +F R W+
Sbjct: 324 WGTRVLATCPCCGNADETVLHVLCDCRPASQVWI-----RLVPSDWITNFFSFDDCRDWV 488
Query: 1675 FGHLQNRDNTL--------FCAMAWNLWKWRNSFVFEQ 1704
F +L R N + F W++W WRN +FE+
Sbjct: 489 FKNLSKRSNGVSKFKWQPTFMTTCWHMWTWRNKAIFEE 602
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 78.2 bits (191), Expect = 3e-14
Identities = 42/116 (36%), Positives = 65/116 (55%)
Frame = +1
Query: 1188 QRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFC 1247
++GLRQGDPLSPYLF+LC L+ ++ + ++V++ I+HL FADD LLF
Sbjct: 7 EKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLFA 186
Query: 1248 QASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRAS 1303
+A+ + I L + ASG VN EKS + +S+ V + + + + I S
Sbjct: 187 RANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEKEMICQQIAIKTGS 354
Score = 31.6 bits (70), Expect(2) = 0.024
Identities = 16/55 (29%), Positives = 28/55 (50%)
Frame = +3
Query: 1527 WNFDRLYTLLPQEIREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQFLNED 1581
WN D ++ +I + + S++ ED+I+W D IYS SA+ L ++
Sbjct: 435 WNRDLIFHSFNNYAAHQIIKIPL-SMRQPEDKIIWHWEKDGIYSVRSAHHALCDE 596
Score = 25.8 bits (55), Expect(2) = 0.024
Identities = 9/18 (50%), Positives = 10/18 (55%)
Frame = +2
Query: 1589 WKLIWKASAPEKVRFFLW 1606
WK IW A V+ FLW
Sbjct: 644 WKAIWNTPASRSVQNFLW 697
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 73.2 bits (178), Expect = 9e-13
Identities = 38/94 (40%), Positives = 57/94 (60%)
Frame = +3
Query: 1187 LQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLF 1246
+QRGL+QGDPL+P+LF+L E ++ ++N V ++ V + G +SHL +ADD L
Sbjct: 18 VQRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCI 197
Query: 1247 CQASKDQLRLIKHTLADFCEASGMKVNLEKSRML 1280
+ D L +K L F ASG+KVN KS ++
Sbjct: 198 GMPTVDNLWTLKALLQGFEMASGLKVNFHKSSLI 299
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 68.2 bits (165), Expect = 3e-11
Identities = 54/216 (25%), Positives = 84/216 (38%), Gaps = 21/216 (9%)
Frame = -3
Query: 1454 LTEELRPGGSYVWRGILKARDSVSDGFGPRLGAG-SSSLWYSNWLGTGKIASRIPFVHIT 1512
L L SY WR I A+ + G +G G ++++W + P H +
Sbjct: 762 LNAPLGSWASYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHSS 583
Query: 1513 DTTLTVADIWRQGAW---------NFDRLYTLLPQEIREEISSVQIPSIQTGEDRILWRE 1563
+G N + + ++ P+ R +I S+ P GED W
Sbjct: 582 RAY*APTLYGYEGCRSDDPMRRERNANLINSIFPEGTRRKILSIH-PQGPIGEDSYSWEY 406
Query: 1564 ASDDIYSASSAYQF-------LNEDGLPET----GFWKLIWKASAPEKVRFFLWLVGRDA 1612
+ YS S Y N+ G + ++ +WK + KVR FLW ++
Sbjct: 405 SKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFLWRCISNS 226
Query: 1613 LPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCP 1648
LPT A H++ SC RCG E H+L +CP
Sbjct: 225 LPTAANMRSRHISKDGSCSRCGMESETVNHILFQCP 118
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 42.0 bits (97), Expect(3) = 2e-09
Identities = 20/46 (43%), Positives = 28/46 (60%)
Frame = +2
Query: 1175 VLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEG 1220
VL N + RGL+QGD LSPY+F++C+E L+ I + E G
Sbjct: 128 VLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERG 265
Score = 36.2 bits (82), Expect(3) = 2e-09
Identities = 24/90 (26%), Positives = 45/90 (49%)
Frame = +3
Query: 1237 LFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGL 1296
L F L + + +++K+ L + E SG ++L KS + S+ V + ++ +
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 1297 VGISRASNLGKYLGLPLLQGRVKRDHFATI 1326
+G+ KYLGLP + GR + F++I
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTFSSI 548
Score = 23.1 bits (48), Expect(3) = 2e-09
Identities = 9/27 (33%), Positives = 16/27 (58%)
Frame = +1
Query: 1135 LEKAYDRVNWDFLRATLYDFGFPPRTV 1161
+ K Y+RV+ D+L+ + GF R +
Sbjct: 7 ISKVYNRVD*DYLKEIMIKMGFNNRWI 87
>BG586862
Length = 804
Score = 54.7 bits (130), Expect = 3e-07
Identities = 43/164 (26%), Positives = 75/164 (45%), Gaps = 5/164 (3%)
Frame = -1
Query: 1592 IWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRC--PG 1649
+W + + FLW + +ALP + H+ + + C RC + +E +H+ C
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEVTQ 475
Query: 1650 SVWVRGQLGRLLPWISDSATFRHWLFGH-LQNRDNTLFC--AMAWNLWKWRNSFVFEQVP 1706
W QLG + S F W+ L+N + T+ A+ +++W RN VFE +
Sbjct: 474 KEWFGSQLG-INFHSSGVLHFHDWITNFILKNDEETIIALTALLYSIWHARNQKVFENID 298
Query: 1707 WQPGDVMRRIHLDTVEFHRLAQGTQRSNPIASDGINSVMLYTDG 1750
PGDV+ + ++ ++AQ + P S+ I S L+ G
Sbjct: 297 -VPGDVVIQRASSSLHSFKMAQVSDSVLP--SNAIPSYSLWGIG 175
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 50.8 bits (120), Expect = 5e-06
Identities = 42/171 (24%), Positives = 64/171 (36%), Gaps = 15/171 (8%)
Frame = +3
Query: 1560 LWREASDDIYSASSAYQFLN----------EDGLPETGFWKLIWKASAPEKVRFFLWLVG 1609
+W IYS S Y L ET WK IW + + LW +
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRIL 182
Query: 1610 RDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCPGS--VWVRGQLGRLLPWISDS 1667
D+LP + + + C RC + E H+ CP S VW L + +
Sbjct: 183 NDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNLCINFDNL-PN 359
Query: 1668 ATFRHWLFGHLQNRDNTL---FCAMAWNLWKWRNSFVFEQVPWQPGDVMRR 1715
F + L+ + +D + A+ +NLW RN V E D+++R
Sbjct: 360 PNFIN*LYEAIL*KDECITI*IAAIIYNLWHARNLSVLEDQTILEMDIIQR 512
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 49.7 bits (117), Expect = 1e-05
Identities = 43/186 (23%), Positives = 73/186 (39%), Gaps = 21/186 (11%)
Frame = -2
Query: 1483 RLGAGSSSL-WYSNWLGTGKIASRIPFVHITDTTLTVA--DIWRQGA--------WNFDR 1531
++G G SS W W + + P L + D+W A W
Sbjct: 703 KVGDGRSSFFWKDAWDSSVPLRESFPRAFFPYRLLKMGCGDLWDMNAEGVRWRLYWRRLE 524
Query: 1532 LYTLLPQEIREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQFLN-----EDGLP-- 1584
L+ + + E + ++ ++ D +W+ + ++S +S Y L ED L
Sbjct: 523 LFEWEKERLLELLGRLEGVVLRYWADIWVWKPDKEGVFSVNSCYFLLQNLRLLEDRLSYE 344
Query: 1585 ETGFWKLIWKASAPEKVRFFLWLVGRDALPTN---AKRHRHHLATTASCIRCGAVVEDAE 1641
E ++ +WK+ AP KV F W + D +PT KR + + C+ CG E
Sbjct: 343 EEVIFRELWKSKAPAKVLAFSWTLFLDRIPTMVNLGKRRLLRVEDSKRCVFCGCQDETVV 164
Query: 1642 HVLRRC 1647
H+ C
Sbjct: 163 HLFLHC 146
>TC82093
Length = 569
Score = 48.1 bits (113), Expect = 3e-05
Identities = 23/47 (48%), Positives = 32/47 (67%)
Frame = -1
Query: 887 ILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLE 933
+L Q EMLW+ KS+ NWV++G RNT FFH + V RN+ GL ++
Sbjct: 476 LLSQNEMLWY*KSQ*NWVKYGSRNTWFFHNEDV--N*RNQTSGLIVD 342
>AW980456
Length = 779
Score = 48.1 bits (113), Expect = 3e-05
Identities = 24/69 (34%), Positives = 34/69 (48%), Gaps = 5/69 (7%)
Frame = -2
Query: 1557 DRILWREASDDIYSASSAYQFLNED-----GLPETGFWKLIWKASAPEKVRFFLWLVGRD 1611
DR++W++ Y SAY+F E+ L G W IWK P KV+ +W + R
Sbjct: 208 DRLIWKDEKHGKYYVKSAYRFCVEELFDSSYLHRPGNWSGIWKLKVPPKVQNLVWRMCRG 29
Query: 1612 ALPTNAKRH 1620
LPT + H
Sbjct: 28 CLPTRIRLH 2
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 43.9 bits (102), Expect = 6e-04
Identities = 47/172 (27%), Positives = 71/172 (40%), Gaps = 16/172 (9%)
Frame = +2
Query: 1519 ADIWRQG------AWNFDRLYTLLPQEIREEISSVQIPSIQTGEDRILWREASDDI--YS 1570
AD+ R G AW + R + +E E + + WR D + YS
Sbjct: 380 ADMGRLGWTVDGRAWEWTRRLFVWEEECVRECCILLNNFVLQDNVNDKWRWLLDPVNGYS 559
Query: 1571 ASSAYQFLNEDG-LPETGFWKLIWKASAPEKVRFFLWLVGRDALPT--NAKRHRHHLATT 1627
Y+++ G + + +W P KV F+W + R+ LPT N LAT
Sbjct: 560 VKVFYRYITSTGHISDRSLVDDVWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVLLATN 739
Query: 1628 ASCIRCGAV-VEDAEHVLRRCP--GSVW--VRGQLGRLLPWISDSATFRHWL 1674
A+C+ CG V E H+ C S+W VR LG +P +S H++
Sbjct: 740 AACV-CGCVDSESTTHLFLHCNVFCSLWSLVRNWLG--IPSMSSGELRTHFI 886
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.336 0.145 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,976,365
Number of Sequences: 36976
Number of extensions: 1042461
Number of successful extensions: 7584
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 4074
Number of HSP's successfully gapped in prelim test: 363
Number of HSP's that attempted gapping in prelim test: 3313
Number of HSP's gapped (non-prelim): 4798
length of query: 1904
length of database: 9,014,727
effective HSP length: 110
effective length of query: 1794
effective length of database: 4,947,367
effective search space: 8875576398
effective search space used: 8875576398
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0228a.4


