

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.12
(414 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
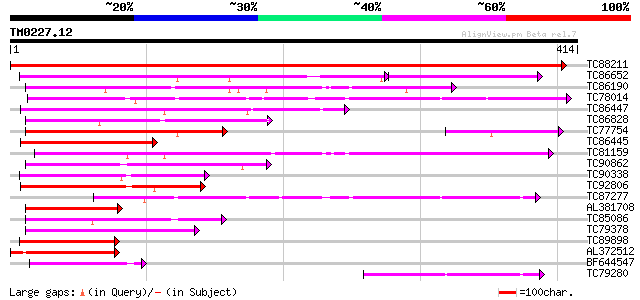
Score E
Sequences producing significant alignments: (bits) Value
TC88211 similar to PIR|E96534|E96534 Hypothetical protein [impor... 672 0.0
TC86652 similar to GP|19911585|dbj|BAB86896. syringolide-induced... 156 1e-53
TC86190 weakly similar to PIR|T08454|T08454 hypothetical protein... 151 5e-37
TC78014 similar to GP|17529090|gb|AAL38755.1 unknown protein {Ar... 147 6e-36
TC86447 weakly similar to GP|19911585|dbj|BAB86896. syringolide-... 141 4e-34
TC86828 similar to PIR|T08454|T08454 hypothetical protein F22O6.... 134 5e-32
TC77754 similar to GP|19911585|dbj|BAB86896. syringolide-induced... 134 5e-32
TC86445 weakly similar to GP|19911585|dbj|BAB86896. syringolide-... 125 2e-29
TC81159 weakly similar to PIR|T45588|T45588 arm repeat containin... 116 1e-26
TC90862 similar to GP|21740496|emb|CAD40820. OSJNBa0006B20.11 {O... 110 1e-24
TC90338 similar to GP|10178069|dbj|BAB11433. emb|CAB89350.1~gene... 105 2e-23
TC92806 similar to GP|10178069|dbj|BAB11433. emb|CAB89350.1~gene... 102 3e-22
TC87277 similar to PIR|T45588|T45588 arm repeat containing prote... 100 1e-21
AL381708 similar to PIR|E96734|E96 unknown protein F23N20.1 [imp... 97 1e-20
TC85086 similar to GP|4510376|gb|AAD21464.1| unknown protein {Ar... 90 2e-18
TC79378 similar to PIR|T00518|T00518 hypothetical protein At2g23... 85 6e-17
TC89898 similar to GP|19715649|gb|AAL91644.1 AT3g07360/F21O3_7 {... 83 2e-16
AL372512 similar to GP|21536926|gb unknown {Arabidopsis thaliana... 83 2e-16
BF644547 similar to GP|14149112|dbj bg55 {Bruguiera gymnorrhiza}... 81 8e-16
TC79280 similar to GP|21592960|gb|AAM64910.1 unknown {Arabidopsi... 78 7e-15
>TC88211 similar to PIR|E96534|E96534 Hypothetical protein [imported] -
Arabidopsis thaliana, partial (67%)
Length = 1478
Score = 672 bits (1734), Expect = 0.0
Identities = 335/406 (82%), Positives = 372/406 (91%)
Frame = +3
Query: 1 MRGCLEPLDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRA 60
MRGCLEPLDLG+QIPYH+RCPISLELMRDPVTV TGQTYDR+SIESWVNTGNTTCPVTR
Sbjct: 117 MRGCLEPLDLGVQIPYHFRCPISLELMRDPVTVSTGQTYDRNSIESWVNTGNTTCPVTRT 296
Query: 61 PLTEFTLIPNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHH 120
LT+FT IPNHTLRRLIQ+WCV+NRAFGV+RIPTPKQPA+ ALVRSLLNQISS SA T
Sbjct: 297 NLTDFTFIPNHTLRRLIQDWCVSNRAFGVQRIPTPKQPADAALVRSLLNQISSHSAPTQL 476
Query: 121 RLNSLRRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESD 180
RLNSLRRLR L RDS+YNRSLI+S +VR I+L I N DEL +ESLAL+V+FPL+ES+
Sbjct: 477 RLNSLRRLRSLTRDSEYNRSLISSLNVRNIILPILFNNGLDELKNESLALIVLFPLSESE 656
Query: 181 CAAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVV 240
C ++A+D DKI YL+ LLSH S DVRVNSAALIE++VAGTHS ++R QVS+V+GI+DGVV
Sbjct: 657 CTSLASDSDKINYLTSLLSHDSFDVRVNSAALIEIIVAGTHSPEIRLQVSNVDGIYDGVV 836
Query: 241 EILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALAT 300
EIL++PISYPRALK+G+KALFALCLVKQ+R RAV+AGAP VLIDRLADFEKCDAERALAT
Sbjct: 837 EILKNPISYPRALKIGIKALFALCLVKQTRHRAVSAGAPVVLIDRLADFEKCDAERALAT 1016
Query: 301 VELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEAVAA 360
VELLCRVPAGCA FAGHALTVPMLVKIILKISDRATEYAAGAL++LCSESER Q E VAA
Sbjct: 1017VELLCRVPAGCASFAGHALTVPMLVKIILKISDRATEYAAGALMALCSESERCQRETVAA 1196
Query: 361 GVLTQLLLLVQSDCTERAKRKAQLLLKLLRDSWPQDSIGNSDDFAC 406
GVLTQLLLLVQSDCTERAKRKAQLLLKLLRDSWPQDS+GNSDDFAC
Sbjct: 1197GVLTQLLLLVQSDCTERAKRKAQLLLKLLRDSWPQDSVGNSDDFAC 1334
>TC86652 similar to GP|19911585|dbj|BAB86896. syringolide-induced protein
13-1-1 {Glycine max}, partial (78%)
Length = 1540
Score = 156 bits (395), Expect(2) = 1e-53
Identities = 98/286 (34%), Positives = 159/286 (55%), Gaps = 16/286 (5%)
Frame = +1
Query: 8 LDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTL 67
L + I IP H+RCP++L+LM+DPVT+ TG TYDR SIE W +GN +CPVT+ LT F +
Sbjct: 145 LQVEIAIPTHFRCPVTLDLMKDPVTLSTGITYDRDSIEKWFESGNNSCPVTKTELTSFDI 324
Query: 68 IPNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHR--LNSL 125
+PNH+LRR+IQ+WCV +R++GVERIPTP+ P V ++I SA+ +
Sbjct: 325 VPNHSLRRMIQDWCVQHRSYGVERIPTPRIPVTRYEVTDTCSRILSAAQQGDESKCFELV 504
Query: 126 RRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRS------SDELSHESLALLV-MFPLAE 178
R+++ ++S+ N+ +I S+ V ++ ++FD+ S + L E L +L M PL+E
Sbjct: 505 RKIKGWGKESERNKKVIVSNGVSLVLAKVFDSFSRGLIEKNVVLLEEILEVLTWMRPLSE 684
Query: 179 SDCAAIAADLDKIGYLSVLLS-HQSLDVRVNSAALI-EMVVAGTHSADLRSQVSSVEGIH 236
+ + L L+ Q + R N++ L+ EM V ++ +EGI
Sbjct: 685 ESRFVFLGSSNSLSCLVWFLNDQQKISTRQNASLLLKEMNV---------ESLAKIEGIV 837
Query: 237 DGVVEILRSPISYPRA-LKVGVKALFALCLVKQSR----QRAVAAG 277
+ +V +++ + A K + +F L +S+ +R VA G
Sbjct: 838 ESLVNMVKVNVEIGSASTKACLSTIFHLVYSSKSKKVILERFVAIG 975
Score = 71.6 bits (174), Expect(2) = 1e-53
Identities = 38/114 (33%), Positives = 67/114 (58%), Gaps = 1/114 (0%)
Frame = +2
Query: 277 GAPAVLIDRLADFEKCDAERALATVELLCRVPAGCAEFAGHALTVPMLVKIILKISDRAT 336
G ++L++ L D EK E+AL + LC G +ALT+P+++K +L++S+ ++
Sbjct: 974 GLVSILLEILVDAEKGVCEKALDVLNCLCDSKNGVQIAKSNALTLPLVIKKLLRVSELSS 1153
Query: 337 EYAAGALLSLCSESER-LQMEAVAAGVLTQLLLLVQSDCTERAKRKAQLLLKLL 389
+ + +C ++E + +EA+ G+ +LL+L+Q C E K KA LLKLL
Sbjct: 1154 SFVVSIVYKICDKAEEGILIEAIQLGMFQKLLVLLQVGCAESTKEKATELLKLL 1315
>TC86190 weakly similar to PIR|T08454|T08454 hypothetical protein F22O6.170
- Arabidopsis thaliana, partial (60%)
Length = 1159
Score = 151 bits (381), Expect = 5e-37
Identities = 111/335 (33%), Positives = 175/335 (52%), Gaps = 20/335 (5%)
Frame = +1
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWV-NTGNTTCPVTRAPLTEFTLI-- 68
+++P + CPISLE+M+DPVTV TG TYDR SIE W+ + N TCPVT+ L+++T +
Sbjct: 124 VEVPSFFVCPISLEIMKDPVTVSTGITYDRESIEKWLFSKKNMTCPVTKQLLSDYTDLTT 303
Query: 69 PNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRL 128
PNHTLRRLIQ WC N + G+ERIPTPK P + L+ + S +S + +L L+RL
Sbjct: 304 PNHTLRRLIQAWCTLNASQGIERIPTPKPPINKTQITKLIKEASHSSLTV--QLKCLKRL 477
Query: 129 RQLARDSDYNRSLIASHDVRRIVLRIFDNRS--------SDELSH-----ESLALLVMFP 175
R +A S+ N+ I + I N S S LS E+L++L
Sbjct: 478 RSMASGSETNKRCIEDAGAVEFIASIVINNSCNIDFVNNSGSLSETNTVDEALSILHNLH 657
Query: 176 LAESDCAAIAA--DLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVE 233
++E+ + A + + I L+ ++ + R + L++ + L Q +E
Sbjct: 658 VSEAGLKKLLAFRNGEFIESLTKVMQKGFFESRAYAVFLLKSMSEVAEPVQLLHQ--KIE 831
Query: 234 GIHDGVVEILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLAD--FEK 291
+ +V++L+ IS + K ++ L ALC ++R + V AG VLI+ L + ++
Sbjct: 832 -LFVELVQVLKDQIS-QKVSKAALQTLIALCPWGRNRIKGVEAGTVPVLIELLLENCKDR 1005
Query: 292 CDAERALATVELLCRVPAGCAEFAGHALTVPMLVK 326
E L +E LC+ G AE HA + ++ K
Sbjct: 1006KPNEMMLVLLENLCQCAEGRAELLRHAAGLAVVSK 1110
>TC78014 similar to GP|17529090|gb|AAL38755.1 unknown protein {Arabidopsis
thaliana}, partial (74%)
Length = 2305
Score = 147 bits (372), Expect = 6e-36
Identities = 122/408 (29%), Positives = 197/408 (47%), Gaps = 11/408 (2%)
Frame = +3
Query: 14 IPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHTL 73
IP +RCPISLELM+DPV V TGQTY+RS I+ W++ G+ TCP T+ L L PN+ L
Sbjct: 984 IPDDFRCPISLELMKDPVIVSTGQTYERSCIQKWLDAGHRTCPKTQQTLLHTALTPNYVL 1163
Query: 74 RRLIQEWCVANRAFGVE-----------RIPTPKQPAEPALVRSLLNQISSASASTHHRL 122
+ LI WC +N GVE + T + +++LL++++S
Sbjct: 1164 KSLIGLWCDSN---GVELPKKQGSCRTKKSGTSLSDCDKTAIKALLDKLTSNDIEQQRA- 1331
Query: 123 NSLRRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCA 182
+ LR LA+ + NR IA +++ + + H ALL + + ES+
Sbjct: 1332 -AAGELRLLAKRNADNRVCIAEAGAIPLLVDLLSSTDPRTQEHADTALLNL-SINESNKG 1505
Query: 183 AIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVEI 242
I + I + +L + S++ R N+AA + + D G ++++
Sbjct: 1506 TI-VNAGAIPDIVDVLKNGSMEARENAAATLFSLSV----LDENKVAIGAAGAIPALIKL 1670
Query: 243 LRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALATVE 302
L PR K A+F LC+ + ++ RAV AG A LI + D + ALA +
Sbjct: 1671 LCE--GTPRGKKDAATAIFNLCIYQGNKARAVKAGIVAPLIRFMKDAGGGMVDEALAILT 1844
Query: 303 LLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEAVAAGV 362
+L G G A +P+LV++I S R E AA L +C+ + LQ++
Sbjct: 1845 ILAGHHEGRTAI-GQAEPIPILVEVIRTGSPRNRENAAAVLWLVCT-GDLLQLKLAKEHG 2018
Query: 363 LTQLLLLVQSDCTERAKRKAQLLLKLLRDSWPQDSIGNSDDFACSQLV 410
+ L + + T+RAKRKA +L+LL+ +DS+ N+ C+ ++
Sbjct: 2019 AEEALQGLSENGTDRAKRKAGSILELLQRIEGEDSLQNT*PLECNFII 2162
>TC86447 weakly similar to GP|19911585|dbj|BAB86896. syringolide-induced
protein 13-1-1 {Glycine max}, partial (36%)
Length = 1223
Score = 141 bits (356), Expect = 4e-34
Identities = 77/249 (30%), Positives = 135/249 (53%), Gaps = 9/249 (3%)
Frame = +1
Query: 9 DLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLI 68
++ + +P +RCPI+L+LM+DPVT+ TG TYDR S+E W N GN TCP+T + F +I
Sbjct: 337 NMEVVVPNQFRCPITLDLMKDPVTLSTGITYDRESVERWFNEGNYTCPLTNQVVRNFDMI 516
Query: 69 PNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQI--SSASASTHHRLNSLR 126
PNH+LR +IQ+WCV NR GVERIPTP+ P P V LL Q+ S+ + + ++
Sbjct: 517 PNHSLRIMIQDWCVENRQNGVERIPTPRIPISPIDVSELLFQVKESAKGLDQYGCIGLVQ 696
Query: 127 RLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLV-------MFPLAES 179
++ + + +S+ N+ I + + FD ++D + + L V MFPL +
Sbjct: 697 KMEKWSNESERNKKCIVENGATSALALAFDAFANDSIEKNDIVLEVILSALNWMFPL-QL 873
Query: 180 DCAAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGV 239
+ + + L HQ + + + ++ +++ + + + +EG+++ +
Sbjct: 874 EAQKSLGSKASLHCMIWFLKHQDVKGKEKAIIALKEILSFGDEKHVEA-LMEIEGVNELL 1050
Query: 240 VEILRSPIS 248
+E + IS
Sbjct: 1051IEFINKRIS 1077
>TC86828 similar to PIR|T08454|T08454 hypothetical protein F22O6.170 -
Arabidopsis thaliana, partial (23%)
Length = 826
Score = 134 bits (338), Expect = 5e-32
Identities = 79/187 (42%), Positives = 106/187 (56%), Gaps = 6/187 (3%)
Frame = +3
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWV-NTGNTTCPVTRAPLTE-----F 65
I++ + CPISL+LM+DPVTV TG TYDR SIE W+ ++ N TCPVT+ LT
Sbjct: 66 IEVTPFFLCPISLQLMKDPVTVSTGITYDRESIEKWLFSSENKTCPVTKQLLTHDANHLI 245
Query: 66 TLIPNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSL 125
L PNHTLRRLIQ WC N ++G+ERIPTPK P L+ LL + AS S H + +L
Sbjct: 246 ILTPNHTLRRLIQAWCTMNSSYGIERIPTPKPPTTKILIEKLLKE---ASDSPHLLIQTL 416
Query: 126 RRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAAIA 185
++L+ +A +S+ NR I S + I ++ S S L+ + D A
Sbjct: 417 KKLKTIASESETNRRCIESAGAVEFLASIVTQNNTSCSSSCSATELIEASFDDDDVEGFA 596
Query: 186 ADLDKIG 192
D KIG
Sbjct: 597 FDF-KIG 614
>TC77754 similar to GP|19911585|dbj|BAB86896. syringolide-induced protein
13-1-1 {Glycine max}, partial (85%)
Length = 1577
Score = 134 bits (338), Expect = 5e-32
Identities = 61/150 (40%), Positives = 97/150 (64%), Gaps = 2/150 (1%)
Frame = +1
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNH 71
I IP ++RCP+SL+LM+DPVT+ TG TYDR SI+ W+ GN TCPVT L+ F + PNH
Sbjct: 190 ITIPTNFRCPVSLDLMKDPVTLSTGITYDRFSIDKWIEAGNKTCPVTNQKLSTFEITPNH 369
Query: 72 TLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHR--LNSLRRLR 129
T+R++IQ WCV N ++G+ERIPTP+ P V + ++ S + + + + +++
Sbjct: 370 TIRKMIQSWCVENSSYGIERIPTPRIPVSGYEVNEVCTRLLSGCRNLDEKKCVEFVGKIK 549
Query: 130 QLARDSDYNRSLIASHDVRRIVLRIFDNRS 159
R+S+ N+ +I + V ++ +FD+ S
Sbjct: 550 IWWRESERNKRVIIGNGVSSVLATVFDSFS 639
Score = 50.8 bits (120), Expect = 9e-07
Identities = 29/89 (32%), Positives = 51/89 (56%), Gaps = 3/89 (3%)
Frame = +3
Query: 319 LTVPMLVKIILKISDRATEYAAGALLSLCSES--ERLQMEAVAAGVLTQLLLLVQSDCTE 376
L +P+++K +L++S A+ +A G + + E ER+ +EA+ G +LL+++Q C E
Sbjct: 1128 LALPLVIKKLLRVSPLASNFAVGIVRKILCEKKEERVLIEAIQLGAFQKLLVMLQVGCEE 1307
Query: 377 RAKRKAQLLLKLLRDSWPQ-DSIGNSDDF 404
+ K LLKLL + + + NS DF
Sbjct: 1308 KTKENTTELLKLLNGYRSKAECVDNSLDF 1394
>TC86445 weakly similar to GP|19911585|dbj|BAB86896. syringolide-induced
protein 13-1-1 {Glycine max}, partial (22%)
Length = 588
Score = 125 bits (315), Expect = 2e-29
Identities = 56/100 (56%), Positives = 72/100 (72%)
Frame = +2
Query: 9 DLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLI 68
++ + +P +RCPI+LELM+DPVT+ TG TYDR S+E W N GN TCP+T + F +I
Sbjct: 284 NMEVVVPNQFRCPITLELMKDPVTLSTGITYDRESVERWFNEGNYTCPLTNQVVRNFDMI 463
Query: 69 PNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLL 108
PNH+LR +IQ+WCV NR GVERIPTP+ P P V LL
Sbjct: 464 PNHSLRIMIQDWCVENRQNGVERIPTPRIPISPLDVSELL 583
>TC81159 weakly similar to PIR|T45588|T45588 arm repeat containing protein
homolog - Arabidopsis thaliana, partial (12%)
Length = 1476
Score = 116 bits (291), Expect = 1e-26
Identities = 112/395 (28%), Positives = 187/395 (46%), Gaps = 16/395 (4%)
Frame = +1
Query: 19 RCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHTLRRLIQ 78
RCPISLELM DPVT+ TG TYDRSSI W +GN+TCP T L L+PN LRRLIQ
Sbjct: 1 RCPISLELMSDPVTIETGHTYDRSSILKWFRSGNSTCPKTGKSLGSIELVPNLVLRRLIQ 180
Query: 79 EWCVAN---------RAFGVERIPTPKQPAEPALVRSLLNQI--SSASASTHHRLNSLRR 127
++C N R+ + R P A + L + S + + + ++
Sbjct: 181 QYCNVNGIPFADSSRRSRDITRTVEPGSVAAEGAMTLLAGFLCRSLDNGNVEQKNHAAFE 360
Query: 128 LRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAAIAA- 186
+R L + S ++RS + ++L + + S + ALL + +S +
Sbjct: 361 VRVLTKTSIFSRSCFVESGLVPLLLLLLASSDSSAQENAIAALLNLSKYIKSRSEMVENW 540
Query: 187 DLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVEILRSP 246
L+ I + VL +++ + ++AA++ + + A+L + E I ++ +++
Sbjct: 541 GLEMI--VGVLNKGINIEAKQHAAAVLFYLASNPEHANLIGE--EPEAI-PSLISLIKD- 702
Query: 247 ISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCD-AERALATVELLC 305
R++K G+ A+F L ++ +R +AA A +L++ L EK D +LA + L
Sbjct: 703 -DNKRSVKNGLVAIFGLLKNHENHKRILAAQAIPLLVNILKASEKEDLVTDSLAILATLA 879
Query: 306 RVPAGCAEFAGH-ALTVPMLVKIILKISDR-ATEYAAGALLSL-CSESERLQMEAVAAGV 362
G +E AL V + V + R E+ LLSL + E + V +
Sbjct: 880 EKSDGTSEILRFGALHVAVEVMSSSSTTSRLGKEHCVSLLLSLSINGGENVIAHLVKSSS 1059
Query: 363 LTQLLLLVQSDCTERAKRKAQLLLKLLRDSWPQDS 397
L + L S+ T RA +KA L+++L D + + S
Sbjct: 1060LMESLYSQLSEGTSRASKKASSLIRVLHDFYERRS 1164
>TC90862 similar to GP|21740496|emb|CAD40820. OSJNBa0006B20.11 {Oryza
sativa}, partial (14%)
Length = 1090
Score = 110 bits (274), Expect = 1e-24
Identities = 63/189 (33%), Positives = 98/189 (51%), Gaps = 9/189 (4%)
Frame = +3
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNH 71
I IP+ +RCPISL+L+ DPVT+ TGQTYDRSSIE W++ GN TCPVT L + + +PNH
Sbjct: 510 ITIPHLFRCPISLDLLEDPVTLTTGQTYDRSSIEKWISAGNFTCPVTMQKLHDLSFVPNH 689
Query: 72 TLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRLRQL 131
TLR LI +W G + P+ A + SL + + S ++ +L SL ++ L
Sbjct: 690 TLRHLIDQWL----QLGAQFEPSSNNSATIDYLASLKHTLESHDSTLETKLKSLEKISVL 857
Query: 132 ARD-SDYNRSLIASHDVRRIVLRIFDNRSSDELSHESL--------ALLVMFPLAESDCA 182
+ + + +S ++L + S +LS + +L + P+ +
Sbjct: 858 SDEYCSFRKSCFQQLSFLPLLLELVFGLSDSQLSENHMEFVEIALSCILKLLPIVNLEPL 1037
Query: 183 AIAADLDKI 191
I D K+
Sbjct: 1038NIIKDESKL 1064
>TC90338 similar to GP|10178069|dbj|BAB11433.
emb|CAB89350.1~gene_id:MUB3.18~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (18%)
Length = 734
Score = 105 bits (263), Expect = 2e-23
Identities = 56/141 (39%), Positives = 85/141 (59%), Gaps = 2/141 (1%)
Frame = +1
Query: 8 LDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTL 67
+DL I +P ++CPISL++M+ PV++CTG TYDRSSI+ W++ GN TCP T L
Sbjct: 76 VDLCITVPTFFKCPISLDVMKSPVSLCTGVTYDRSSIQRWLDDGNNTCPATMQILPTKDF 255
Query: 68 IPNHTLRRLIQEW--CVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSL 125
+PN TL LIQ W V +R VE + +P + L++++ + SS T +R +
Sbjct: 256 VPNRTLHSLIQIWTDSVHHR---VEPVVSPSVLSNQQLLQTITDLASSGLNRTLNRFGLI 426
Query: 126 RRLRQLARDSDYNRSLIASHD 146
++ A+DSD NR+ +A D
Sbjct: 427 MKVIHFAQDSDQNRTFLAKLD 489
>TC92806 similar to GP|10178069|dbj|BAB11433.
emb|CAB89350.1~gene_id:MUB3.18~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (23%)
Length = 565
Score = 102 bits (254), Expect = 3e-22
Identities = 54/138 (39%), Positives = 83/138 (60%), Gaps = 3/138 (2%)
Frame = +2
Query: 9 DLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLI 68
DL I +P +RCPISL++M+ PV++CTG TYDRSSI+ W++ GN TCP T L +
Sbjct: 86 DLCITVPSLFRCPISLDVMKSPVSLCTGVTYDRSSIQRWLDNGNNTCPATMQILQTKDFV 265
Query: 69 PNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALV---RSLLNQISSASASTHHRLNSL 125
PN TL+RLIQ W + R R+ +P+ P V L+ I+ + + ++ +SL
Sbjct: 266 PNRTLQRLIQIWSDSVR----HRVDSPESPLSTESVDRRDQLIVAITDFESGSENQFDSL 433
Query: 126 RRLRQLARDSDYNRSLIA 143
++ + A+DS+ N +A
Sbjct: 434 VKIVRFAKDSEENCVFLA 487
>TC87277 similar to PIR|T45588|T45588 arm repeat containing protein homolog
- Arabidopsis thaliana, partial (48%)
Length = 1393
Score = 100 bits (248), Expect = 1e-21
Identities = 101/335 (30%), Positives = 154/335 (45%), Gaps = 9/335 (2%)
Frame = +1
Query: 62 LTEFTLIPNHTLRRLIQEWCVANRAFGVERIPTPKQ--------PAEPALVRSLLNQISS 113
LT L PN+ LR LI++WC AN +R T + PAE + + SL+ +++S
Sbjct: 1 LTSTVLTPNYVLRSLIEQWCEANGIEPPKRPSTSEPSKSASACTPAERSKIESLIQKLTS 180
Query: 114 ASASTHHRLNSLRRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVM 173
++ +R LA+ + NR IA +++ + S H ALL +
Sbjct: 181 GGPEDQR--SAAGEIRLLAKRNADNRVAIAEAGAIPLLVGLLSVPDSRTQEHAVTALLNL 354
Query: 174 FPLAESDCAAIAADLDKIGYLSVLLSHQSLDVRVNSAA-LIEMVVAGTHSADLRSQVSSV 232
+ ES+ +I + G + VL S++ R N+AA L + V + + S
Sbjct: 355 -SIYESNKGSIVSSGAVPGIVHVL-KRGSMEARENAAATLFSLSVIDENKVTIGSS---- 516
Query: 233 EGIHDGVVEILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKC 292
G +V +L R K ALF LC+ + ++ +AV AG L+ L +
Sbjct: 517 -GAISPLVTLLSEGTQ--RGKKDAATALFNLCIYQGNKGKAVRAGVIPTLMRLLTEPSGG 687
Query: 293 DAERALATVELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESER 352
+ ALA + +L P G A G A VP+LVK I S R E AA L+ LCS +
Sbjct: 688 MVDEALAILAILASHPEGKAAI-GAAEAVPVLVKFIGNGSPRNKENAAAVLVHLCSGDRQ 864
Query: 353 LQMEAVAAGVLTQLLLLVQSDCTERAKRKAQLLLK 387
+A GV+ LL L Q+ T+R KRKA L++
Sbjct: 865 YIAQAQELGVMAPLLELAQNG-TDRGKRKATQLIE 966
>AL381708 similar to PIR|E96734|E96 unknown protein F23N20.1 [imported] -
Arabidopsis thaliana, partial (18%)
Length = 482
Score = 97.1 bits (240), Expect = 1e-20
Identities = 42/71 (59%), Positives = 52/71 (73%)
Frame = +3
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNH 71
I IP + CPISLELMRDPV V TGQTY+RS I+ W++ GNTTCP T+ L TL PN+
Sbjct: 132 IVIPEDFLCPISLELMRDPVIVATGQTYERSYIQRWIDCGNTTCPKTQQKLQHLTLTPNY 311
Query: 72 TLRRLIQEWCV 82
LR L+ +WC+
Sbjct: 312 VLRSLVSQWCI 344
>TC85086 similar to GP|4510376|gb|AAD21464.1| unknown protein {Arabidopsis
thaliana}, partial (15%)
Length = 656
Score = 89.7 bits (221), Expect = 2e-18
Identities = 52/151 (34%), Positives = 83/151 (54%), Gaps = 4/151 (2%)
Frame = +3
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGN-TTCPVTR--APLTEFTLI 68
I+IP ++ CPIS ++M +PVT TG TYDR SIE W+ CPVT P + L
Sbjct: 123 IEIPPYFVCPISFQIMEEPVTTVTGITYDRKSIEKWLMKAKICVCPVTNQSLPRSSEYLT 302
Query: 69 PNHTLRRLIQEWCVANRAFGVE-RIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRR 127
PNHTL+RLI+ W ++N A V+ +I +PK P ++ L+ + + S+ +
Sbjct: 303 PNHTLQRLIKAWILSNEAKVVDNQIQSPKSPLNRIHLQKLVKNLELPNCFQ----ASMEK 470
Query: 128 LRQLARDSDYNRSLIASHDVRRIVLRIFDNR 158
+ +LA+ SD NR + V + ++ + +
Sbjct: 471 ILELAKQSDRNRICMVEVGVTKAMVMVIKKK 563
>TC79378 similar to PIR|T00518|T00518 hypothetical protein At2g23140
[imported] - Arabidopsis thaliana, partial (36%)
Length = 2133
Score = 84.7 bits (208), Expect = 6e-17
Identities = 44/127 (34%), Positives = 68/127 (52%)
Frame = +1
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNH 71
+ +P + CP+SLELM DPV V +GQTY+R+ I++W++ G T CP T L LIPN+
Sbjct: 826 VPVPPDFCCPLSLELMTDPVIVASGQTYERAFIKNWIDLGLTVCPKTHQTLAHTNLIPNY 1005
Query: 72 TLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRLRQL 131
T++ LI WC +N V+ I + L + + + S H ++L +
Sbjct: 1006TVKALIANWCESNNVKLVDPIKSTTLNQASILHGYMESGTTRESPVFPHSRSNLPSSPES 1185
Query: 132 ARDSDYN 138
AR +N
Sbjct: 1186ARSRSFN 1206
>TC89898 similar to GP|19715649|gb|AAL91644.1 AT3g07360/F21O3_7 {Arabidopsis
thaliana}, partial (15%)
Length = 816
Score = 83.2 bits (204), Expect = 2e-16
Identities = 34/73 (46%), Positives = 47/73 (63%)
Frame = +1
Query: 8 LDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTL 67
++ + P ++CP+S E MRDPV V +GQTYDR I+ W N GN TCP T L+ L
Sbjct: 379 INKNVTFPDEFKCPLSKEFMRDPVIVASGQTYDRPFIQRWFNAGNRTCPQTHQVLSHTLL 558
Query: 68 IPNHTLRRLIQEW 80
PNH +R +I++W
Sbjct: 559 TPNHLIREMIEKW 597
>AL372512 similar to GP|21536926|gb unknown {Arabidopsis thaliana}, partial
(34%)
Length = 462
Score = 82.8 bits (203), Expect = 2e-16
Identities = 39/80 (48%), Positives = 55/80 (68%)
Frame = +1
Query: 1 MRGCLEPLDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRA 60
+R ++ L+L ++P + CPISLE M+DPVT+CTGQTY+R++I W N G+ TCP T
Sbjct: 13 LRKMIQELEL-CEVPSVFICPISLEPMQDPVTLCTGQTYERNNILKWFNMGHFTCPTTMQ 189
Query: 61 PLTEFTLIPNHTLRRLIQEW 80
L + ++ PN TL RLI W
Sbjct: 190ELWDDSITPNTTLYRLIYTW 249
>BF644547 similar to GP|14149112|dbj bg55 {Bruguiera gymnorrhiza}, partial
(8%)
Length = 681
Score = 80.9 bits (198), Expect = 8e-16
Identities = 37/86 (43%), Positives = 48/86 (55%)
Frame = +1
Query: 15 PYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHTLR 74
P Y CPISL LM DPV + +G+TY+R I+ W + GN CP T+ L + PN L+
Sbjct: 376 PEEYACPISLRLMYDPVVIASGETYERMWIQKWFDEGNVICPKTKKKLLHLAMTPNVALK 555
Query: 75 RLIQEWCVANRAFGVERIPTPKQPAE 100
LI +WC N IP P + AE
Sbjct: 556 ELISKWCKTNDV----SIPNPSRQAE 621
>TC79280 similar to GP|21592960|gb|AAM64910.1 unknown {Arabidopsis
thaliana}, partial (46%)
Length = 809
Score = 77.8 bits (190), Expect = 7e-15
Identities = 49/132 (37%), Positives = 74/132 (55%)
Frame = +2
Query: 259 ALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALATVELLCRVPAGCAEFAGHA 318
AL+ LC VK+++ RAV AG VL++ +ADFE +++ + +L VP
Sbjct: 104 ALYTLCSVKENKMRAVKAGIMKVLVELMADFESNMVDKSAYVLSVLVSVPEAKVALVEEG 283
Query: 319 LTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEAVAAGVLTQLLLLVQSDCTERA 378
VP+LV+I+ S R E AA LL +C +S ++ G + L+ L QS T RA
Sbjct: 284 -GVPVLVEIVEVGSQRQKEIAAVILLQICEDSVAVRSMVAREGAIPPLVALTQSG-TNRA 457
Query: 379 KRKAQLLLKLLR 390
K+KA+ L++LLR
Sbjct: 458 KQKAEKLIELLR 493
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,078,278
Number of Sequences: 36976
Number of extensions: 153520
Number of successful extensions: 715
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 692
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 693
length of query: 414
length of database: 9,014,727
effective HSP length: 99
effective length of query: 315
effective length of database: 5,354,103
effective search space: 1686542445
effective search space used: 1686542445
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0227.12


