

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0221.11
(142 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
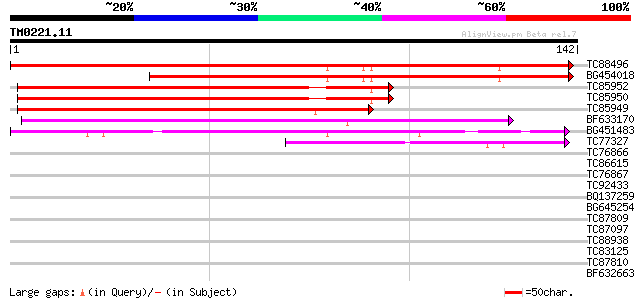
Score E
Sequences producing significant alignments: (bits) Value
TC88496 similar to GP|1079652|gb|AAD09209.1| late embryogenesis ... 211 6e-56
BG454018 similar to GP|1079652|gb| late embryogenesis abundant p... 153 3e-38
TC85952 similar to GP|11612179|gb|AAG37442.1 seed maturation pro... 122 6e-29
TC85950 similar to GP|11612179|gb|AAG37442.1 seed maturation pro... 122 6e-29
TC85949 similar to GP|10334835|gb|AAG15413.1 seed maturation pro... 119 4e-28
BF633170 similar to GP|4836407|gb|A seed maturation protein PM29... 47 2e-06
BG451483 weakly similar to PIR|PN0109|PN01 keratin-like protein ... 42 8e-05
TC77327 homologue to GP|16224254|gb|AAL15651.1 dehydrin-like pro... 40 2e-04
TC76866 similar to GP|2909420|emb|CAA12026.1 LEA protein {Cicer ... 36 0.006
TC86615 weakly similar to GP|8778409|gb|AAF79417.1| F16A14.5 {Ar... 34 0.017
TC76867 similar to GP|2909420|emb|CAA12026.1 LEA protein {Cicer ... 34 0.022
TC92433 weakly similar to GP|21592859|gb|AAM64809.1 unknown {Ara... 32 0.065
BQ137259 homologue to GP|15387793|emb hypothetical predicted pro... 31 0.14
BG645254 weakly similar to GP|21436677|em putative histidine pro... 30 0.25
TC87809 similar to PIR|A24403|UMMS period clock protein - mouse ... 30 0.32
TC87097 similar to GP|14334540|gb|AAK59678.1 putative protein {A... 30 0.32
TC88938 similar to PIR|F85436|F85436 hypothetical protein AT4g36... 30 0.32
TC83125 similar to PIR|T05841|T05841 spliceosome-associated prot... 30 0.42
TC87810 similar to GP|21592752|gb|AAM64701.1 unknown {Arabidopsi... 30 0.42
BF632663 similar to GP|21689765|gb putative arsA-like protein hA... 29 0.55
>TC88496 similar to GP|1079652|gb|AAD09209.1| late embryogenesis abundant
protein {Glycine soja}, partial (96%)
Length = 874
Score = 211 bits (538), Expect = 6e-56
Identities = 119/176 (67%), Positives = 127/176 (71%), Gaps = 35/176 (19%)
Frame = +1
Query: 1 MQGAKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVS 60
MQGAKKA ET+KETAANIGASAKSG EKTKAT+QEK EK+TA DPVQKEMAT KKEAK++
Sbjct: 91 MQGAKKAGETIKETAANIGASAKSGMEKTKATLQEKTEKMTARDPVQKEMATHKKEAKMN 270
Query: 61 QAELDKLAAREHNAATKQ-SAAAAAHMG-----TG-TGTGTAAYSTTGAHGQPMGAHQTS 113
QAELDKLAAREHNAA KQ + AAA HMG TG TGTGTA YSTTG +G P GAHQ S
Sbjct: 271 QAELDKLAAREHNAAVKQTTTAAAGHMGQPHHTTGTTGTGTATYSTTGNYGHPTGAHQMS 450
Query: 114 ALPGHGTG----------------------------HNTRVGGNPNATGYGTGGTY 141
A+PGHGTG HNTRVGGNPNATGY TGGTY
Sbjct: 451 AMPGHGTGQPTGHVVDGVVGSHPIGTNRGTDGTATAHNTRVGGNPNATGYTTGGTY 618
>BG454018 similar to GP|1079652|gb| late embryogenesis abundant protein
{Glycine soja}, partial (75%)
Length = 586
Score = 153 bits (386), Expect = 3e-38
Identities = 87/141 (61%), Positives = 94/141 (65%), Gaps = 35/141 (24%)
Frame = +3
Query: 36 KAEKVTAGDPVQKEMATQKKEAKVSQAELDKLAAREHNAATKQ-SAAAAAHMG-----TG 89
+ EK+TA DPVQKEMAT KKEAK++QAELDKLAAREHNAA KQ + AAA HMG TG
Sbjct: 105 QTEKMTARDPVQKEMATHKKEAKMNQAELDKLAAREHNAAVKQTTTAAAGHMGQPHHTTG 284
Query: 90 -TGTGTAAYSTTGAHGQPMGAHQTSALPGHGTG--------------------------- 121
TGTGTA YSTTG +G P GAHQ SA+PGHGTG
Sbjct: 285 TTGTGTATYSTTGNYGHPTGAHQMSAMPGHGTGQPTGHVVDGVVGSHPIGTNRGTDGTAT 464
Query: 122 -HNTRVGGNPNATGYGTGGTY 141
HNTRVGGNPNATGY TGGTY
Sbjct: 465 AHNTRVGGNPNATGYTTGGTY 527
>TC85952 similar to GP|11612179|gb|AAG37442.1 seed maturation protein LEA 4
{Glycine tabacina}, partial (49%)
Length = 530
Score = 122 bits (305), Expect = 6e-29
Identities = 69/98 (70%), Positives = 77/98 (78%), Gaps = 4/98 (4%)
Frame = +3
Query: 3 GAKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQA 62
GAKK E++KETAANIGASAKSG EKTKAT QEK EK+TA DP+QKEMATQKKE +V+QA
Sbjct: 75 GAKKTGESIKETAANIGASAKSGMEKTKATFQEKTEKMTAHDPLQKEMATQKKEERVNQA 254
Query: 63 ELDKLAAREHNAATKQSAAAAAHMGTG----TGTGTAA 96
ELDK AAREHNAA A+A +G G TGTG AA
Sbjct: 255 ELDKEAAREHNAA----ASAGHQLGVGGHHTTGTGGAA 356
>TC85950 similar to GP|11612179|gb|AAG37442.1 seed maturation protein LEA 4
{Glycine tabacina}, partial (49%)
Length = 807
Score = 122 bits (305), Expect = 6e-29
Identities = 69/98 (70%), Positives = 77/98 (78%), Gaps = 4/98 (4%)
Frame = +2
Query: 3 GAKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQA 62
GAKK E++KETAANIGASAKSG EKTKAT QEK EK+TA DP+QKEMATQKKE +V+QA
Sbjct: 92 GAKKTGESIKETAANIGASAKSGMEKTKATFQEKTEKMTAHDPLQKEMATQKKEERVNQA 271
Query: 63 ELDKLAAREHNAATKQSAAAAAHMGTG----TGTGTAA 96
ELDK AAREHNAA A+A +G G TGTG AA
Sbjct: 272 ELDKEAAREHNAA----ASAGHQLGVGGHHTTGTGGAA 373
>TC85949 similar to GP|10334835|gb|AAG15413.1 seed maturation protein
{Glycine tomentella}, partial (44%)
Length = 653
Score = 119 bits (298), Expect = 4e-28
Identities = 64/91 (70%), Positives = 72/91 (78%), Gaps = 2/91 (2%)
Frame = +1
Query: 3 GAKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQA 62
GAKK E++KETAANIGASAKSG EKTKAT+QEK EK+TA DP+QKEMATQKKE +V+QA
Sbjct: 97 GAKKTGESIKETAANIGASAKSGMEKTKATLQEKTEKMTAHDPLQKEMATQKKEERVNQA 276
Query: 63 ELDKLAAREHNAA--TKQSAAAAAHMGTGTG 91
ELDK AAREHNAA T H +GTG
Sbjct: 277 ELDKEAAREHNAAATTGHQLGQGGHHTSGTG 369
>BF633170 similar to GP|4836407|gb|A seed maturation protein PM29 {Glycine
max}, partial (63%)
Length = 640
Score = 47.4 bits (111), Expect = 2e-06
Identities = 34/126 (26%), Positives = 56/126 (43%), Gaps = 3/126 (2%)
Frame = +2
Query: 4 AKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQAE 63
A+ ++ E N ++AK + KA + EKAEK A +K +A ++ +AK +A+
Sbjct: 23 ARGEMQSSMEKLKNKASAAKEQADIYKAKIDEKAEKRMARTKEEKVIAHERAKAKEHKAK 202
Query: 64 LDKLAAREHNAATKQSAAAA---AHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGT 120
++ A+ +AA K H G G G +P+G HQ A+ G+
Sbjct: 203 MELHEAKARHAAEKLHTKKPHYYGHHGPVEGVQQPQPQVVGTKNEPVGVHQPEAVVGNQY 382
Query: 121 GHNTRV 126
N V
Sbjct: 383 QENEPV 400
>BG451483 weakly similar to PIR|PN0109|PN01 keratin-like protein - rat
(fragment), partial (15%)
Length = 673
Score = 42.0 bits (97), Expect = 8e-05
Identities = 44/146 (30%), Positives = 64/146 (43%), Gaps = 6/146 (4%)
Frame = +3
Query: 1 MQGAKKATETMKETAANI-GASA--KSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEA 57
M A+K +T E + GA+A ++G ++ KA K G+ + K +A K++
Sbjct: 99 MGKAEKHGDTTPEAHNQLQGANAHKQAGAQEGKAGAGHA--KAGIGEKLSKNVAETKEKL 272
Query: 58 KVSQAELDKLAAREHNAATKQ--SAAAAAHMGTGTGTGTAAYSTTG-AHGQPMGAHQTSA 114
+ + AA+EH+ A ++ AAAA H A S AH TS+
Sbjct: 273 HIGNKHGEATAAKEHHKANEKEHKAAAAGHAEKVADHADAGASRVDKAHSHETANRGTSS 452
Query: 115 LPGHGTGHNTRVGGNPNATGYGTGGT 140
H TGH T V G TG TG T
Sbjct: 453 ---HTTGHTTGVAG--GTTGLTTGAT 515
>TC77327 homologue to GP|16224254|gb|AAL15651.1 dehydrin-like protein
{Medicago sativa}, partial (71%)
Length = 1330
Score = 40.4 bits (93), Expect = 2e-04
Identities = 27/75 (36%), Positives = 37/75 (49%), Gaps = 4/75 (5%)
Frame = +2
Query: 70 REHNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGH---GTGH-NTR 125
+ H Q+ ++ GTGTG GT S G H +G + T+ G G+G+ NT
Sbjct: 254 QHHGVGVDQTTGFGSNTGTGTGYGTHTGSG-GTHTGGVGGYGTTTEYGSTNTGSGYGNTD 430
Query: 126 VGGNPNATGYGTGGT 140
+GG TG GTG T
Sbjct: 431 IGGTGYGTGTGTGTT 475
Score = 37.4 bits (85), Expect = 0.002
Identities = 26/65 (40%), Positives = 30/65 (46%), Gaps = 10/65 (15%)
Frame = +2
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTR----------VGGNPNATGYG 136
GTGTGTGT Y TG G +G GTGH+ R G + NA+ YG
Sbjct: 449 GTGTGTGTTGYGATGG-GTGVGY--------GGTGHDNRGVMDKIKEKIPGTDQNASTYG 601
Query: 137 TGGTY 141
TG Y
Sbjct: 602 TGTGY 616
Score = 34.3 bits (77), Expect = 0.017
Identities = 24/68 (35%), Positives = 27/68 (39%), Gaps = 18/68 (26%)
Frame = +2
Query: 87 GTGTGTGTAAYSTTGA----HGQP----------MGAHQTSALPGHGTGHNTR----VGG 128
GTGTG GT Y +G HG+ G Q + G GTGH T G
Sbjct: 734 GTGTGYGTTGYGASGVGHQQHGEKGVMDKIKEKIPGTEQNTYGTGTGTGHGTTGYGSTGT 913
Query: 129 NPNATGYG 136
TGYG
Sbjct: 914 GHGTTGYG 937
Score = 33.9 bits (76), Expect = 0.022
Identities = 24/85 (28%), Positives = 31/85 (36%), Gaps = 11/85 (12%)
Frame = +2
Query: 53 QKKEAKVSQAELDKLAAREHNA---ATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGA 109
Q E V +K+ E N T + TGTG GT Y HG+ G
Sbjct: 791 QHGEKGVMDKIKEKIPGTEQNTYGTGTGTGHGTTGYGSTGTGHGTTGYGDEQHHGEKKGI 970
Query: 110 HQ--------TSALPGHGTGHNTRV 126
+ T + GHG GH T +
Sbjct: 971 MEKIKEKLPGTGSCTGHGQGH*TMI 1045
Score = 31.2 bits (69), Expect = 0.14
Identities = 23/77 (29%), Positives = 28/77 (35%), Gaps = 2/77 (2%)
Frame = +2
Query: 65 DKLAAREHNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQT--SALPGHGTGH 122
+K+ + NA+T GTGTG GT G G +PG
Sbjct: 560 EKIPGTDQNAST---------YGTGTGYGTTGIGHQQHGGDNRGVMDKIKEKIPGTDQNQ 712
Query: 123 NTRVGGNPNATGYGTGG 139
T G TGYGT G
Sbjct: 713 YTHGTGTGTGTGYGTTG 763
Score = 25.8 bits (55), Expect = 6.1
Identities = 18/55 (32%), Positives = 23/55 (41%), Gaps = 16/55 (29%)
Frame = +2
Query: 103 HGQPMGAHQTSALPGHGTGHN-----------TRVGGNP-NATGYGT----GGTY 141
+G P+ + GHG GH+ T G N TGYGT GGT+
Sbjct: 188 YGNPISGGGMTGATGHGHGHHQQHHGVGVDQTTGFGSNTGTGTGYGTHTGSGGTH 352
>TC76866 similar to GP|2909420|emb|CAA12026.1 LEA protein {Cicer arietinum},
partial (85%)
Length = 781
Score = 35.8 bits (81), Expect = 0.006
Identities = 32/105 (30%), Positives = 44/105 (41%), Gaps = 9/105 (8%)
Frame = +2
Query: 6 KATETMKETA-------ANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAK 58
KA ETM T NIG A++ KEK + T Q EK Q A ++K +
Sbjct: 98 KAGETMGRTEEKSNQMMGNIGDKAQAAKEKVQQTAQAAKEKTG-----QTAQAAKEKTQE 262
Query: 59 VSQAELDKL--AAREHNAATKQSAAAAAHMGTGTGTGTAAYSTTG 101
+QA DK AA+ T+ + A G G T + +G
Sbjct: 263 TAQAAKDKTQQAAQATKDKTQDTTGQARDKGYEMGQATKETAQSG 397
>TC86615 weakly similar to GP|8778409|gb|AAF79417.1| F16A14.5 {Arabidopsis
thaliana}, partial (31%)
Length = 1278
Score = 34.3 bits (77), Expect = 0.017
Identities = 26/55 (47%), Positives = 27/55 (48%), Gaps = 1/55 (1%)
Frame = +2
Query: 87 GTGTGTGTAAYSTTG-AHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTGGT 140
GTGTGTGT + TG G MG T G GTG T G TG GTG T
Sbjct: 662 GTGTGTGTGTGTPTGTGTGTGMG---TPTGTGTGTGIGTGTG-----TGTGTGTT 802
Score = 27.7 bits (60), Expect = 1.6
Identities = 12/26 (46%), Positives = 14/26 (53%)
Frame = +2
Query: 76 TKQSAAAAAHMGTGTGTGTAAYSTTG 101
T +GTGTGTGT +TTG
Sbjct: 731 TPTGTGTGTGIGTGTGTGTGTGTTTG 808
>TC76867 similar to GP|2909420|emb|CAA12026.1 LEA protein {Cicer arietinum},
partial (85%)
Length = 828
Score = 33.9 bits (76), Expect = 0.022
Identities = 31/105 (29%), Positives = 43/105 (40%), Gaps = 9/105 (8%)
Frame = +1
Query: 6 KATETMKETA-------ANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAK 58
KA ETM T NIG A++ KEK + T Q EK Q A ++K +
Sbjct: 118 KAGETMGRTEEKSNQMMGNIGDKAQAAKEKVQQTAQAAKEKTG-----QTAQAAKEKTQE 282
Query: 59 VSQAELDKL--AAREHNAATKQSAAAAAHMGTGTGTGTAAYSTTG 101
+QA DK A+ T+ + A G G T + +G
Sbjct: 283 TAQAVKDKTQQTAQATKDKTQDTTGQARDKGYEMGQATKETAQSG 417
>TC92433 weakly similar to GP|21592859|gb|AAM64809.1 unknown {Arabidopsis
thaliana}, partial (53%)
Length = 686
Score = 32.3 bits (72), Expect = 0.065
Identities = 22/68 (32%), Positives = 27/68 (39%)
Frame = +3
Query: 73 NAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNA 132
NA T + + + GTGTGTGT + TG G GTG G+PN
Sbjct: 447 NAGTPSTGSPGSTPGTGTGTGTGTGTGTGT--------------GTGTG---STAGSPNV 575
Query: 133 TGYGTGGT 140
G T
Sbjct: 576 IGISPSST 599
>BQ137259 homologue to GP|15387793|emb hypothetical predicted protein
LM15-1.83 unknown function {Leishmania major}, partial
(3%)
Length = 1050
Score = 31.2 bits (69), Expect = 0.14
Identities = 22/97 (22%), Positives = 42/97 (42%)
Frame = +2
Query: 2 QGAKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQ 61
+G++ ET G S G++ T + A + ++E ++E++ +
Sbjct: 635 RGSRAPRETASRA*TTRGHSRTQGRQTRTRTATDHAAMKRQRE--ERERRAYRRESR-ER 805
Query: 62 AELDKLAAREHNAATKQSAAAAAHMGTGTGTGTAAYS 98
+ +L REHN + + A H GTGT A++
Sbjct: 806 EQRRELRNREHNEQLRVAQDRARHRGTGTAGDRRAHA 916
>BG645254 weakly similar to GP|21436677|em putative histidine protein kinase
{Rhodobacter sphaeroides}, partial (2%)
Length = 553
Score = 30.4 bits (67), Expect = 0.25
Identities = 32/113 (28%), Positives = 44/113 (38%), Gaps = 18/113 (15%)
Frame = +2
Query: 48 KEMATQKKEAKVSQAELDKLAAREHNAATKQSAA--------AAAH------MGTGTGTG 93
K MA K A++ A AAR +AA + +AA A+AH M T G
Sbjct: 113 KRMAKAKLRARLGAAAGVSAAARHVHAAVRHAAAARSSAVLDASAHAAWEGLMNTVPGPN 292
Query: 94 TAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVG----GNPNATGYGTGGTYN 142
AA + T + T+ + G NT++ G P G GG N
Sbjct: 293 FAADAQTNGEMHWLRQWWTAVVARQSPGVNTQMAELMVGAPGIAGGAAGGAAN 451
>TC87809 similar to PIR|A24403|UMMS period clock protein - mouse (fragment),
partial (5%)
Length = 670
Score = 30.0 bits (66), Expect = 0.32
Identities = 23/64 (35%), Positives = 27/64 (41%)
Frame = +1
Query: 76 TKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGY 135
T S + TGTGTGT TT + G TS G GTG +T GN +
Sbjct: 448 TSGSGTSTTSPSTGTGTGT-GMGTTPSTSTGTGT-GTSTGTGMGTGTSTGATGNTPYSTT 621
Query: 136 GTGG 139
GG
Sbjct: 622 AXGG 633
>TC87097 similar to GP|14334540|gb|AAK59678.1 putative protein {Arabidopsis
thaliana}, partial (62%)
Length = 1234
Score = 30.0 bits (66), Expect = 0.32
Identities = 19/62 (30%), Positives = 31/62 (49%)
Frame = +1
Query: 7 ATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQAELDK 66
AT M+ GAS+ S T+ ++++ AGD Q+ A++KKE K + E +
Sbjct: 214 ATRRMRRRTVASGASSSSAPPATQQESGDESDNEAAGDE-QEAKASKKKELKRQEREARR 390
Query: 67 LA 68
A
Sbjct: 391 QA 396
>TC88938 similar to PIR|F85436|F85436 hypothetical protein AT4g36970
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1701
Score = 30.0 bits (66), Expect = 0.32
Identities = 26/109 (23%), Positives = 44/109 (39%), Gaps = 12/109 (11%)
Frame = +2
Query: 12 KETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQK----------EMATQKKEAKVSQ 61
KET+ + S + KT + + + K+TA + +QK EM +KK A
Sbjct: 968 KETSTRSSSWEISERSKTVSKAKREEAKITAWENLQKAKAEAAIQKLEMKLEKKRASSMD 1147
Query: 62 AELDKLAAREHNAATKQSAAAA--AHMGTGTGTGTAAYSTTGAHGQPMG 108
++KL + A +S+ + AH T ++ G G G
Sbjct: 1148 KIMNKLKFAQKKAQEMRSSVSVDQAHQVARTSHKVMSFRRAGQMGSLSG 1294
Score = 26.2 bits (56), Expect = 4.7
Identities = 11/27 (40%), Positives = 15/27 (54%)
Frame = +2
Query: 75 ATKQSAAAAAHMGTGTGTGTAAYSTTG 101
A K+S + GTGT TGT + + G
Sbjct: 83 AVKKSMVQSRKSGTGTATGTGTFPSPG 163
>TC83125 similar to PIR|T05841|T05841 spliceosome-associated protein homolog
F17L22.120 - Arabidopsis thaliana, partial (24%)
Length = 722
Score = 29.6 bits (65), Expect = 0.42
Identities = 23/88 (26%), Positives = 40/88 (45%), Gaps = 11/88 (12%)
Frame = +2
Query: 4 AKKATETMKETAANIGASAKSGKEKT-------KATVQEKAEKVTAGDPVQKEMATQKKE 56
AKK + ++ A+NIG + + KE T + ++ EKV + + +E ++
Sbjct: 128 AKKNNKASEQPASNIGEESDNAKENTDPKQVFEQVEIEYVPEKVDLYEGMDEEFRKIFEK 307
Query: 57 AKVSQA----ELDKLAAREHNAATKQSA 80
++ E DK E AATK+ A
Sbjct: 308 FSFTEVAASEETDKKDVAEETAATKKKA 391
>TC87810 similar to GP|21592752|gb|AAM64701.1 unknown {Arabidopsis
thaliana}, partial (52%)
Length = 1094
Score = 29.6 bits (65), Expect = 0.42
Identities = 22/67 (32%), Positives = 30/67 (43%)
Frame = +3
Query: 73 NAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNA 132
+A+ ++ + GTGTGTG +T G G TS G GTG +T GN
Sbjct: 483 SASGSGTSTTSPSTGTGTGTGMGTTPSTST-GTGTG---TSTGTGMGTGTSTGATGNTPY 650
Query: 133 TGYGTGG 139
+ GG
Sbjct: 651 STTAPGG 671
Score = 28.9 bits (63), Expect = 0.72
Identities = 21/65 (32%), Positives = 28/65 (42%), Gaps = 4/65 (6%)
Frame = +3
Query: 76 TKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSA----LPGHGTGHNTRVGGNPN 131
T S + GT TGTG ++TGA G + T+A L G G+G G N
Sbjct: 552 TTPSTSTGTGTGTSTGTGMGTGTSTGATGNT--PYSTTAPGGVLGGIGSGMGPSGAGGMN 725
Query: 132 ATGYG 136
+G
Sbjct: 726 DDSHG 740
>BF632663 similar to GP|21689765|gb putative arsA-like protein hASNA-I
{Arabidopsis thaliana}, partial (41%)
Length = 594
Score = 29.3 bits (64), Expect = 0.55
Identities = 20/65 (30%), Positives = 24/65 (36%), Gaps = 2/65 (3%)
Frame = +1
Query: 72 HNAATKQSAAAAA--HMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGN 129
H A +AAAA H G GT G H G H H TG + + GG+
Sbjct: 250 HGAHWPHAAAAAVSNHPGKGTQQADEPAGVDGRHDGAGGRHDGRWRGRHATGTHGQAGGD 429
Query: 130 PNATG 134
G
Sbjct: 430 EGCGG 444
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.301 0.116 0.312
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,617,675
Number of Sequences: 36976
Number of extensions: 27000
Number of successful extensions: 225
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 171
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 194
length of query: 142
length of database: 9,014,727
effective HSP length: 87
effective length of query: 55
effective length of database: 5,797,815
effective search space: 318879825
effective search space used: 318879825
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0221.11


