

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
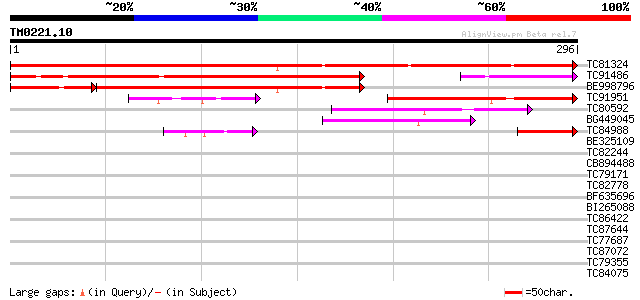
Query= TM0221.10
(296 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81324 similar to GP|21553871|gb|AAM62964.1 unknown {Arabidopsi... 477 e-135
TC91486 similar to GP|5441893|dbj|BAA82391.1 ESTs C99174(E10437)... 254 2e-68
BE998796 similar to GP|5441893|dbj| ESTs C99174(E10437) D22295(C... 217 4e-57
TC91951 similar to GP|5441893|dbj|BAA82391.1 ESTs C99174(E10437)... 150 6e-37
TC80592 similar to GP|19310435|gb|AAL84954.1 F17H15.1/F17H15.1 {... 52 4e-07
BG449045 similar to GP|19310435|gb| F17H15.1/F17H15.1 {Arabidops... 47 7e-06
TC84988 similar to GP|14018368|emb|CAC38358. zinc finger protein... 42 2e-04
BE325109 similar to PIR|T04011|T040 hypothetical protein T5L19.2... 38 0.004
TC82244 similar to GP|22507458|gb|AAH19429.1 Unknown (protein fo... 38 0.004
CB894488 similar to GP|6437560|gb| hypothetical protein {Arabido... 38 0.004
TC79171 similar to GP|19310435|gb|AAL84954.1 F17H15.1/F17H15.1 {... 37 0.012
TC82778 similar to PIR|T04011|T04011 hypothetical protein T5L19.... 35 0.027
BF635696 35 0.027
BI265088 35 0.027
TC86422 similar to PIR|T48439|T48439 probable RNA-binding protei... 35 0.035
TC87644 similar to GP|10177307|dbj|BAB10568. contains similarity... 35 0.045
TC77687 similar to PIR|D84581|D84581 probable CCCH-type zinc fin... 34 0.078
TC87072 similar to PIR|D84581|D84581 probable CCCH-type zinc fin... 34 0.078
TC79355 similar to GP|15081721|gb|AAK82515.1 At1g04990/F13M7_1 {... 33 0.10
TC84075 similar to GP|3738297|gb|AAC63639.1| unknown protein {Ar... 33 0.13
>TC81324 similar to GP|21553871|gb|AAM62964.1 unknown {Arabidopsis
thaliana}, partial (75%)
Length = 1156
Score = 477 bits (1228), Expect = e-135
Identities = 241/304 (79%), Positives = 261/304 (85%), Gaps = 8/304 (2%)
Frame = +1
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHY 60
MD RKRGRPE GFN+NGG KKS+ E+ESLS+GVGSKSKPCTKFFST+GCPFGESCHFLH+
Sbjct: 91 MDMRKRGRPEHGFNNNGGFKKSRHEMESLSTGVGSKSKPCTKFFSTSGCPFGESCHFLHH 270
Query: 61 VPGGYNAVAHMMNLAPSAQAPPRNVAAP-PPPVPNGSTP-AVKTRICNKFNTAEGCKFGD 118
VPGGYNAV+ MMNL P+A PRNV AP PNGS P AVK+RIC+KFNTAEGCKFGD
Sbjct: 271 VPGGYNAVSQMMNLTPAAPPAPRNVPAPRNAHAPNGSAPSAVKSRICSKFNTAEGCKFGD 450
Query: 119 KCHFAHGEWELGKHIAPSFD-----DHRPIGHAPAGRIGG-RMEPPPGPATGFGANATAK 172
KCHFAHGEWELGK +APSFD DHR +G AGR GG RMEPPP PA+ FGANATAK
Sbjct: 451 KCHFAHGEWELGKPVAPSFDDHRHNDHRHMGPPNAGRFGGHRMEPPPVPAS-FGANATAK 627
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMV 232
ISVEASLAGAIIGKGGVNSKQICRQTGAKL+IREHE DPNL+NIEL G+F+QIK+ASNMV
Sbjct: 628 ISVEASLAGAIIGKGGVNSKQICRQTGAKLAIREHE-DPNLKNIELVGTFEQIKDASNMV 804
Query: 233 KDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGPA 292
KDLLLTLQMSAPPKSNQG G G GHHG NN KTKLCENFAKG+CTFG+RCHFAHG
Sbjct: 805 KDLLLTLQMSAPPKSNQGPPGHHGAPGHHG-NNLKTKLCENFAKGSCTFGDRCHFAHGAV 981
Query: 293 ELRK 296
ELRK
Sbjct: 982 ELRK 993
>TC91486 similar to GP|5441893|dbj|BAA82391.1 ESTs C99174(E10437)
D22295(C10709) correspond to a region of the predicted
gene.~Similar to, partial (25%)
Length = 736
Score = 254 bits (650), Expect = 2e-68
Identities = 130/187 (69%), Positives = 146/187 (77%), Gaps = 2/187 (1%)
Frame = +2
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHY 60
MD RKRGR + N G +KK+KQE++S +GVGSK+KPCTKFFS AGCPFGE CHF H+
Sbjct: 194 MDARKRGRVDAALN--GSVKKTKQEMDS--TGVGSKTKPCTKFFSIAGCPFGEGCHFSHH 361
Query: 61 VPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTP-AVKTRICNKFNTAEGCKFGDK 119
VPGGYNAVA MMNL P+A PPRNVAAPPP NGS AVK+RICNKFN+ EGCKFGDK
Sbjct: 362 VPGGYNAVAQMMNLKPAA--PPRNVAAPPPVSNNGSAQHAVKSRICNKFNSLEGCKFGDK 535
Query: 120 CHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPG-PATGFGANATAKISVEAS 178
CHFAHGEWELGK APS DDHR + GR+ GR+E P G T FGAN+TAKI V+AS
Sbjct: 536 CHFAHGEWELGKPYAPSVDDHRLMAPTNVGRMPGRVEAPSGLGGTSFGANSTAKIXVKAS 715
Query: 179 LAGAIIG 185
LAGAIIG
Sbjct: 716 LAGAIIG 736
Score = 40.8 bits (94), Expect = 6e-04
Identities = 21/62 (33%), Positives = 35/62 (55%), Gaps = 1/62 (1%)
Frame = +2
Query: 236 LLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENF-AKGTCTFGERCHFAHGPAEL 294
++ L+ +APP++ A P + + K+++C F + C FG++CHFAHG EL
Sbjct: 392 MMNLKPAAPPRNV--AAPPPVSNNGSAQHAVKSRICNKFNSLEGCKFGDKCHFAHGEWEL 565
Query: 295 RK 296
K
Sbjct: 566 GK 571
Score = 29.6 bits (65), Expect = 1.5
Identities = 13/24 (54%), Positives = 14/24 (58%), Gaps = 1/24 (4%)
Frame = +2
Query: 267 KTKLCENFAK-GTCTFGERCHFAH 289
KTK C F C FGE CHF+H
Sbjct: 287 KTKPCTKFFSIAGCPFGEGCHFSH 358
>BE998796 similar to GP|5441893|dbj| ESTs C99174(E10437) D22295(C10709)
correspond to a region of the predicted gene.~Similar
to, partial (28%)
Length = 609
Score = 217 bits (553), Expect = 4e-57
Identities = 112/148 (75%), Positives = 121/148 (81%), Gaps = 8/148 (5%)
Frame = +1
Query: 46 TAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPP-VPNGSTP-AVKTR 103
T+GCPFGESCHFLH+VPGGYNAV+ MMNL P+A PRNV AP PNGS P AVK+R
Sbjct: 169 TSGCPFGESCHFLHHVPGGYNAVSQMMNLTPAAPPAPRNVPAPRNAHAPNGSAPSAVKSR 348
Query: 104 ICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFD-----DHRPIGHAPAGRIGG-RMEP 157
IC+KFNTAEGCKFGDKCHFAHGEWELGK +APSFD DHR +G AGR GG RMEP
Sbjct: 349 ICSKFNTAEGCKFGDKCHFAHGEWELGKPVAPSFDDHRHNDHRHMGPPNAGRFGGHRMEP 528
Query: 158 PPGPATGFGANATAKISVEASLAGAIIG 185
PP PA+ FGANATAKISVEASLAGAIIG
Sbjct: 529 PPVPAS-FGANATAKISVEASLAGAIIG 609
Score = 46.6 bits (109), Expect = 1e-05
Identities = 23/45 (51%), Positives = 30/45 (66%)
Frame = +2
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFS 45
MD RKRGRPE GFN+NGG KKS+ L L++ + + CT F +
Sbjct: 95 MDMRKRGRPEHGFNNNGGFKKSRHAL--LAAHLVRAATSCTMFLA 223
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/65 (38%), Positives = 36/65 (54%), Gaps = 4/65 (6%)
Frame = +1
Query: 236 LLTLQMSAPPKSNQGGAGGPGGHGHHGS--NNFKTKLCENF--AKGTCTFGERCHFAHGP 291
++ L +APP A H +GS + K+++C F A+G C FG++CHFAHG
Sbjct: 244 MMNLTPAAPPAPRNVPAPR-NAHAPNGSAPSAVKSRICSKFNTAEG-CKFGDKCHFAHGE 417
Query: 292 AELRK 296
EL K
Sbjct: 418 WELGK 432
>TC91951 similar to GP|5441893|dbj|BAA82391.1 ESTs C99174(E10437)
D22295(C10709) correspond to a region of the predicted
gene.~Similar to, partial (23%)
Length = 526
Score = 150 bits (379), Expect = 6e-37
Identities = 75/101 (74%), Positives = 81/101 (79%), Gaps = 2/101 (1%)
Frame = +3
Query: 198 TGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQG--GAGGP 255
TG K SIR+HE+DPNLRNIELEG+FDQI +ASNMVKDLLLTL +SAPPKS G GGP
Sbjct: 3 TGVKFSIRDHETDPNLRNIELEGTFDQIAQASNMVKDLLLTLSVSAPPKSTPGAPAGGGP 182
Query: 256 GGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
G N KTKLCENFAKG+CTFGERCHFAHG AELRK
Sbjct: 183 AAPG----RNIKTKLCENFAKGSCTFGERCHFAHGAAELRK 293
Score = 47.8 bits (112), Expect = 5e-06
Identities = 30/81 (37%), Positives = 42/81 (51%), Gaps = 12/81 (14%)
Frame = +3
Query: 63 GGYNAVAHMMNLAP------SAQAPPRNVAAPPPPVPNGSTPA-----VKTRICNKFNTA 111
G ++ +A N+ S APP++ P P G PA +KT++C F A
Sbjct: 69 GTFDQIAQASNMVKDLLLTLSVSAPPKST----PGAPAGGGPAAPGRNIKTKLCENF--A 230
Query: 112 EG-CKFGDKCHFAHGEWELGK 131
+G C FG++CHFAHG EL K
Sbjct: 231 KGSCTFGERCHFAHGAAELRK 293
Score = 29.6 bits (65), Expect = 1.5
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = +3
Query: 32 GVGSKSKPCTKFFSTAGCPFGESCHFLH 59
G K+K C F + C FGE CHF H
Sbjct: 192 GRNIKTKLCENF-AKGSCTFGERCHFAH 272
>TC80592 similar to GP|19310435|gb|AAL84954.1 F17H15.1/F17H15.1 {Arabidopsis
thaliana}, partial (30%)
Length = 945
Score = 51.6 bits (122), Expect = 4e-07
Identities = 35/108 (32%), Positives = 55/108 (50%), Gaps = 3/108 (2%)
Frame = +1
Query: 169 ATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNLRN--IELEGSFDQI 225
++ KI + G IIGKGG K + Q+GAK+ + R+ ++DPN N +EL G+ D I
Sbjct: 364 SSKKIEIPNGRVGVIIGKGGETIKYLQLQSGAKIQVTRDMDADPNSPNRLVELTGTSDAI 543
Query: 226 KEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCEN 273
A ++K++L A +S G G G++ F K+ N
Sbjct: 544 ATAEKLIKEVL------AEAESGGNGLVTRRMTGQGGADEFSMKIPNN 669
Score = 35.8 bits (81), Expect = 0.020
Identities = 20/60 (33%), Positives = 32/60 (53%), Gaps = 4/60 (6%)
Frame = +2
Query: 181 GAIIGKGGVNSKQICRQTGAKLSIREHESDP----NLRNIELEGSFDQIKEASNMVKDLL 236
G IIGKGG K + TGA++ + P R +++EG+ +QI+ A +V +L
Sbjct: 677 GLIIGKGGETIKSMQATTGARIQVIPLHLPPGDTSTERTLKIEGTSEQIESAKQLVDSIL 856
>BG449045 similar to GP|19310435|gb| F17H15.1/F17H15.1 {Arabidopsis
thaliana}, partial (13%)
Length = 572
Score = 47.4 bits (111), Expect = 7e-06
Identities = 26/83 (31%), Positives = 45/83 (53%), Gaps = 3/83 (3%)
Frame = +1
Query: 164 GFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPN--LRNIELEG 220
G ++ KI + G +IGKGG K + Q+GAK+ + R+ ++DPN R +EL G
Sbjct: 280 GGSYGSSKKIEIPNGRVGVLIGKGGETIKYLQMQSGAKIQVTRDMDADPNSQTRMVELMG 459
Query: 221 SFDQIKEASNMVKDLLLTLQMSA 243
+ D + A ++ ++L + A
Sbjct: 460 TPDAVSSAEKLINEVLAEAEAGA 528
>TC84988 similar to GP|14018368|emb|CAC38358. zinc finger protein {Pisum
sativum}, partial (28%)
Length = 656
Score = 42.4 bits (98), Expect = 2e-04
Identities = 16/31 (51%), Positives = 22/31 (70%)
Frame = +1
Query: 266 FKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
FKT++C F GTC GE C++AHG E+R+
Sbjct: 490 FKTRICTKFRFGTCRNGENCNYAHGADEIRQ 582
Score = 41.2 bits (95), Expect = 5e-04
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 6/55 (10%)
Frame = +1
Query: 81 PPRNVAAPPP----PVPNGSTPAV--KTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
PP + PP P PN T ++ KTRIC KF C+ G+ C++AHG E+
Sbjct: 415 PPSGLMCPPSRMIQPPPNRGTSSIFFKTRICTKFRFGT-CRNGENCNYAHGADEI 576
>BE325109 similar to PIR|T04011|T040 hypothetical protein T5L19.200 -
Arabidopsis thaliana, partial (9%)
Length = 430
Score = 38.1 bits (87), Expect = 0.004
Identities = 24/69 (34%), Positives = 37/69 (52%), Gaps = 4/69 (5%)
Frame = +1
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLS-IREH---ESDPNLRNIELEGSFDQIKE 227
+I V G IIGKGG K + +TGA++ I +H D R +++ G QI+
Sbjct: 19 QIQVPNEKVGLIIGKGGETIKSLQTKTGARIQLIPQHLPEGDDSKERTVQVTGDKRQIEI 198
Query: 228 ASNMVKDLL 236
A M+K++L
Sbjct: 199 AQEMIKEVL 225
>TC82244 similar to GP|22507458|gb|AAH19429.1 Unknown (protein for
MGC:30371) {Mus musculus}, partial (13%)
Length = 847
Score = 38.1 bits (87), Expect = 0.004
Identities = 27/82 (32%), Positives = 36/82 (42%), Gaps = 5/82 (6%)
Frame = +3
Query: 7 GRPEPGFNSNGGIKKSKQELESLSSGVGSK--SKPCTKFFSTAGCPFGESCHFLH--YVP 62
G+ E + SN K+ + E S + K C + T C FGESC F H +VP
Sbjct: 600 GQNEAWYTSNSLAKRPRYETGSTLPIYPQRPGEKDCAHYMLTRTCKFGESCKFDHPIWVP 779
Query: 63 -GGYNAVAHMMNLAPSAQAPPR 83
GG + N+ PS P R
Sbjct: 780 AGGIPXWKEVPNIVPSETLPER 845
Score = 27.3 bits (59), Expect = 7.3
Identities = 15/49 (30%), Positives = 19/49 (38%), Gaps = 5/49 (10%)
Frame = +3
Query: 84 NVAAPPPPVPNGSTPAVKT-----RICNKFNTAEGCKFGDKCHFAHGEW 127
N A P GST + + C + CKFG+ C F H W
Sbjct: 627 NSLAKRPRYETGSTLPIYPQRPGEKDCAHYMLTRTCKFGESCKFDHPIW 773
>CB894488 similar to GP|6437560|gb| hypothetical protein {Arabidopsis
thaliana}, partial (37%)
Length = 830
Score = 38.1 bits (87), Expect = 0.004
Identities = 34/134 (25%), Positives = 47/134 (34%), Gaps = 12/134 (8%)
Frame = +1
Query: 3 FRKRGRPEP-----GFNSNGGIKKSKQELESLSSGVGSKSKP-------CTKFFSTAGCP 50
F + +P+P G +S G+++ +L S G G S P CT + T C
Sbjct: 136 FPRSTQPDPSSQWTGPDSQTGLEEPMWQLGLGSGGSGEDSYPQRPDEVDCTYYLRTGFCG 315
Query: 51 FGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNT 110
FG C F H + A A R V P V +C +
Sbjct: 316 FGSRCRF-----------NHPRDRAAVIGAASRTVGEYP--------ERVGQPVCQYYMR 438
Query: 111 AEGCKFGDKCHFAH 124
CKFG C + H
Sbjct: 439 TRSCKFGASCKYHH 480
Score = 35.4 bits (80), Expect = 0.027
Identities = 24/65 (36%), Positives = 31/65 (46%)
Frame = +1
Query: 38 KPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGST 97
K C+ F T C FG +C F H VP A + APS P V++ PVP+
Sbjct: 556 KECSYFVKTGQCKFGATCKFDHPVP------ASVQIPAPSPVPP---VSSLHVPVPSPLY 708
Query: 98 PAVKT 102
P V+T
Sbjct: 709 PTVQT 723
>TC79171 similar to GP|19310435|gb|AAL84954.1 F17H15.1/F17H15.1 {Arabidopsis
thaliana}, partial (27%)
Length = 1421
Score = 36.6 bits (83), Expect = 0.012
Identities = 25/84 (29%), Positives = 39/84 (45%), Gaps = 4/84 (4%)
Frame = +2
Query: 181 GAIIGKGGVNSKQICRQTGAKLSIREHESDP----NLRNIELEGSFDQIKEASNMVKDLL 236
G IIGKGG K + TGA++ + P R ++++G+ DQI+ A +V +L
Sbjct: 2 GLIIGKGGETIKGMQASTGARIQVIPLHPPPGDTSTERTLKIDGTPDQIESAKQLVNQIL 181
Query: 237 LTLQMSAPPKSNQGGAGGPGGHGH 260
N G +GG G+
Sbjct: 182 ----TGENRLRNSGNSGGYTQQGY 241
>TC82778 similar to PIR|T04011|T04011 hypothetical protein T5L19.200 -
Arabidopsis thaliana, partial (7%)
Length = 784
Score = 35.4 bits (80), Expect = 0.027
Identities = 24/74 (32%), Positives = 40/74 (53%), Gaps = 3/74 (4%)
Frame = +3
Query: 153 GRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDP 211
G+ +P G T T KI V ++ G +IGK G + + +GAK+ I R+ ++DP
Sbjct: 573 GQKQPISGSDT-----TTRKIEVPSNKVGVLIGKSGDTIRYLQYNSGAKIQITRDADADP 737
Query: 212 --NLRNIELEGSFD 223
+ R +EL G+ +
Sbjct: 738 HSSTRPVELIGTLE 779
>BF635696
Length = 545
Score = 35.4 bits (80), Expect = 0.027
Identities = 16/31 (51%), Positives = 19/31 (60%)
Frame = +2
Query: 266 FKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
FKTKLC + G C + C FAHG ELR+
Sbjct: 134 FKTKLCALYQNGRCN-RQNCSFAHGNTELRR 223
>BI265088
Length = 635
Score = 35.4 bits (80), Expect = 0.027
Identities = 16/31 (51%), Positives = 19/31 (60%)
Frame = +3
Query: 266 FKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
FKTKLC + G C + C FAHG ELR+
Sbjct: 135 FKTKLCALYQNGRCN-RQNCSFAHGNTELRR 224
>TC86422 similar to PIR|T48439|T48439 probable RNA-binding protein -
Arabidopsis thaliana, partial (88%)
Length = 1591
Score = 35.0 bits (79), Expect = 0.035
Identities = 25/86 (29%), Positives = 42/86 (48%), Gaps = 4/86 (4%)
Frame = +1
Query: 157 PPPGPATGFGANAT-AKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNL- 213
PP P+ T + V S AG++IGKGG Q+GA++ + R +E P
Sbjct: 214 PPDSPSQDSVEKPTYVRFLVSNSAAGSVIGKGGSTITDFQSQSGARIQLSRNNEFFPGTT 393
Query: 214 -RNIELEGSFDQIKEASNMVKDLLLT 238
R I + G+ +++ A ++ LL+
Sbjct: 394 DRIIMVSGAINEVLRAVELILSKLLS 471
Score = 32.3 bits (72), Expect = 0.23
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 3/63 (4%)
Frame = +1
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIR---EHESDPNLRNIELEGSFDQIKEAS 229
I V G ++G+GG N I + +GAK+ I ++ S R + + GS I+ A
Sbjct: 925 IGVADEHIGLVVGRGGRNISDISQTSGAKIKISDRGDYISGTTDRKVTITGSQRAIRTAE 1104
Query: 230 NMV 232
+M+
Sbjct: 1105SMI 1113
>TC87644 similar to GP|10177307|dbj|BAB10568. contains similarity to zinc
finger protein~gene_id:MDC12.23 {Arabidopsis thaliana},
partial (36%)
Length = 1541
Score = 34.7 bits (78), Expect = 0.045
Identities = 26/88 (29%), Positives = 37/88 (41%), Gaps = 1/88 (1%)
Frame = +1
Query: 40 CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPV-PNGSTP 98
C+ F T C F +C F H N VA + PP N++ P+ P+ S
Sbjct: 958 CSFFIKTGDCKFKSNCKFHH----PKNRVAKL---------PPCNLSDKGLPLRPDQS-- 1092
Query: 99 AVKTRICNKFNTAEGCKFGDKCHFAHGE 126
+C+ ++ CKFG C F H E
Sbjct: 1093 -----VCSHYSRYGICKFGPACRFDHPE 1161
Score = 31.2 bits (69), Expect = 0.50
Identities = 23/106 (21%), Positives = 36/106 (33%)
Frame = +1
Query: 19 IKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSA 78
+++ +E E G C + + GC FG++C + H G+ A +N
Sbjct: 367 VREKVREREEPEENAGQTE--CKYYQRSGGCKFGKACKYNH--SRGFTAPISELNFL--- 525
Query: 79 QAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAH 124
G + R C + CKFG C F H
Sbjct: 526 ----------------GLPIRLGERECPYYMRTGSCKFGSNCRFNH 615
Score = 30.4 bits (67), Expect = 0.86
Identities = 14/50 (28%), Positives = 20/50 (40%)
Frame = +1
Query: 13 FNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVP 62
+N + G EL L + + C + T C FG +C F H P
Sbjct: 475 YNHSRGFTAPISELNFLGLPIRLGERECPYYMRTGSCKFGSNCRFNHPDP 624
>TC77687 similar to PIR|D84581|D84581 probable CCCH-type zinc finger protein
[imported] - Arabidopsis thaliana, partial (35%)
Length = 1096
Score = 33.9 bits (76), Expect = 0.078
Identities = 15/30 (50%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Frame = +3
Query: 265 NFKTKLCENFAK-GTCTFGERCHFAHGPAE 293
N+ C F K G CT G+ CHFAHG E
Sbjct: 315 NYSGNPCPEFRKLGNCTKGDSCHFAHGVFE 404
Score = 33.5 bits (75), Expect = 0.10
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = +3
Query: 105 CNKFNTAEGCKFGDKCHFAHGEWELGKH 132
C +F C GD CHFAHG +E H
Sbjct: 333 CPEFRKLGNCTKGDSCHFAHGVFECWLH 416
Score = 30.0 bits (66), Expect = 1.1
Identities = 11/23 (47%), Positives = 13/23 (55%)
Frame = +3
Query: 39 PCTKFFSTAGCPFGESCHFLHYV 61
PC +F C G+SCHF H V
Sbjct: 330 PCPEFRKLGNCTKGDSCHFAHGV 398
>TC87072 similar to PIR|D84581|D84581 probable CCCH-type zinc finger protein
[imported] - Arabidopsis thaliana, partial (47%)
Length = 1599
Score = 33.9 bits (76), Expect = 0.078
Identities = 13/29 (44%), Positives = 17/29 (57%)
Frame = +3
Query: 265 NFKTKLCENFAKGTCTFGERCHFAHGPAE 293
N+ C +F KG+C G+ C FAHG E
Sbjct: 387 NYSGTSCPDFRKGSCKKGDSCEFAHGVFE 473
Score = 28.9 bits (63), Expect = 2.5
Identities = 11/19 (57%), Positives = 12/19 (62%)
Frame = +3
Query: 114 CKFGDKCHFAHGEWELGKH 132
CK GD C FAHG +E H
Sbjct: 429 CKKGDSCEFAHGVFECWLH 485
>TC79355 similar to GP|15081721|gb|AAK82515.1 At1g04990/F13M7_1 {Arabidopsis
thaliana}, partial (42%)
Length = 1627
Score = 33.5 bits (75), Expect = 0.10
Identities = 27/106 (25%), Positives = 32/106 (29%), Gaps = 8/106 (7%)
Frame = +2
Query: 27 ESLSSGVGSKSK--------PCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSA 78
E+LS G S C F ST C +G C F H
Sbjct: 959 ETLSGGQAMNSSLPDRPEQPDCKYFMSTGTCKYGSDCKFHH------------------- 1081
Query: 79 QAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAH 124
P +A P G IC+ + CKFG C F H
Sbjct: 1082 --PKERIAQTLSINPLGLPMRPGNAICSYYRIYGVCKFGPTCKFDH 1213
Score = 26.9 bits (58), Expect = 9.5
Identities = 15/46 (32%), Positives = 22/46 (47%), Gaps = 5/46 (10%)
Frame = +2
Query: 19 IKKSKQELESLSSGV-----GSKSKPCTKFFSTAGCPFGESCHFLH 59
I+K+ + L LSS + + K C + T C FG +C F H
Sbjct: 527 IQKTGEVLHPLSSIL*VFPCVKEEKSCPYYMRTGSCKFGVACKFHH 664
>TC84075 similar to GP|3738297|gb|AAC63639.1| unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 615
Score = 33.1 bits (74), Expect = 0.13
Identities = 29/104 (27%), Positives = 35/104 (32%), Gaps = 7/104 (6%)
Frame = +3
Query: 28 SLSSGVGSKSKP-------CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQA 80
SL SG G +S P C + T C +G C F H P AVA +
Sbjct: 294 SLGSGGGVESYPERHGVPNCAYYMRTGFCGYGGRCRFNH--PRDRAAVAAAVRATGDY-- 461
Query: 81 PPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAH 124
P + PP C + CKFG C F H
Sbjct: 462 -PERLGEPP---------------CQYYLKTGTCKFGASCKFHH 545
Score = 30.0 bits (66), Expect = 1.1
Identities = 12/26 (46%), Positives = 13/26 (49%)
Frame = +3
Query: 39 PCTKFFSTAGCPFGESCHFLHYVPGG 64
PC + T C FG SC F H GG
Sbjct: 483 PCQYYLKTGTCKFGASCKFHHPKNGG 560
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.135 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,271,258
Number of Sequences: 36976
Number of extensions: 194096
Number of successful extensions: 2858
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 1770
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2703
length of query: 296
length of database: 9,014,727
effective HSP length: 95
effective length of query: 201
effective length of database: 5,502,007
effective search space: 1105903407
effective search space used: 1105903407
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0221.10


