

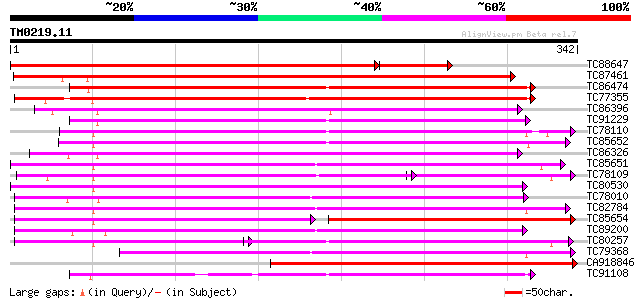
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0219.11
(342 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88647 weakly similar to GP|2688828|gb|AAB88878.1| ethylene-for... 377 e-125
TC87461 weakly similar to GP|2688828|gb|AAB88878.1| ethylene-for... 315 1e-86
TC86474 weakly similar to GP|6016680|gb|AAF01507.1| putative leu... 216 1e-56
TC77355 weakly similar to PIR|T05552|T05552 SRG1 protein-related... 215 2e-56
TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin ... 214 3e-56
TC91229 similar to GP|9294689|dbj|BAB03055.1 gene_id:MHC9.10~sim... 209 9e-55
TC78110 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 203 7e-53
TC85652 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 197 6e-51
TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoantho... 197 6e-51
TC85651 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 196 1e-50
TC78109 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 154 2e-50
TC80530 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabi... 195 2e-50
TC78010 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 194 5e-50
TC82784 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 191 3e-49
TC85654 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 129 1e-47
TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 185 2e-47
TC80257 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 133 3e-47
TC79368 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabi... 180 6e-46
CA918846 weakly similar to GP|21537316|gb ethylene-forming-enzym... 178 3e-45
TC91108 similar to PIR|T05552|T05552 SRG1 protein-related protei... 176 1e-44
>TC88647 weakly similar to GP|2688828|gb|AAB88878.1|
ethylene-forming-enzyme-like dioxygenase {Prunus
armeniaca}, partial (47%)
Length = 978
Score = 377 bits (968), Expect(2) = e-125
Identities = 190/224 (84%), Positives = 205/224 (90%), Gaps = 1/224 (0%)
Frame = +1
Query: 1 MSKSVQEMSMDSDEPPSAYVVERNSFGSK-DSSTLIPIPIIDVSLLSSEDEQGKLRSALS 59
+ +SVQEMSMD DEPPS Y+V N FGSK DSSTLIPIPIIDVSLLSSEDE KLRSALS
Sbjct: 163 LPRSVQEMSMDGDEPPSQYLVNGNVFGSKEDSSTLIPIPIIDVSLLSSEDELEKLRSALS 342
Query: 60 SAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVL 119
SAGCFQAIGH MS++YLDK+REVAK FFALPVEEKQKYARAVNE+EGYGNDR+VS KQVL
Sbjct: 343 SAGCFQAIGHDMSTSYLDKVREVAKQFFALPVEEKQKYARAVNESEGYGNDRIVSDKQVL 522
Query: 120 DWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGS 179
DWSYRL+LRVFPKEK+RLSLWPENPSDF ++L EFSTKVKSMMD+LLR+MARSLNLEEGS
Sbjct: 523 DWSYRLTLRVFPKEKQRLSLWPENPSDFSDTLEEFSTKVKSMMDYLLRSMARSLNLEEGS 702
Query: 180 FLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQ 223
FL QFG+QS AR NFYPPCSRPDLVLGVKPHTDRSGITVLLQ
Sbjct: 703 FLDQFGKQSLFQARINFYPPCSRPDLVLGVKPHTDRSGITVLLQ 834
Score = 88.2 bits (217), Expect(2) = e-125
Identities = 40/44 (90%), Positives = 44/44 (99%)
Frame = +2
Query: 224 DREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSP 267
D+EVEGLQVL+DDKW+NVPTIP+ALVVNLGDQMQIMSNGIFKSP
Sbjct: 836 DKEVEGLQVLIDDKWINVPTIPNALVVNLGDQMQIMSNGIFKSP 967
>TC87461 weakly similar to GP|2688828|gb|AAB88878.1|
ethylene-forming-enzyme-like dioxygenase {Prunus
armeniaca}, partial (62%)
Length = 993
Score = 315 bits (808), Expect = 1e-86
Identities = 159/312 (50%), Positives = 223/312 (70%), Gaps = 9/312 (2%)
Frame = +2
Query: 3 KSVQEMSMDSDEPPSA-YVVERNSFGSKD----SSTLIPIPIIDVSLL----SSEDEQGK 53
KSVQE+S+DS+ PS Y+ + G +D S + + IP++D+ L +S+ E K
Sbjct: 56 KSVQELSLDSENLPSNNYIYKEGGVGFRDALLPSQSDLHIPVVDIGKLISPSTSQQELHK 235
Query: 54 LRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVV 113
L SALSS G FQAI HGM+S L+K+RE++K FF L EEKQKYAR N EGYGND ++
Sbjct: 236 LHSALSSWGLFQAINHGMTSLTLNKVREISKQFFELSKEEKQKYAREPNGIEGYGNDVIL 415
Query: 114 SKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSL 173
S+ Q LDW+ R+ L+V P+ +R L+P+ P+DF ++ +++ ++ + + +LR +A+SL
Sbjct: 416 SENQKLDWTDRVYLKVHPEHQRNFKLFPQKPNDFRNTIEQYTQSLRQLYEIILRAVAKSL 595
Query: 174 NLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL 233
NLEE FL + GE+ ++ R N+YPPC PD VLGVKPH D S IT LLQD+EVEGLQVL
Sbjct: 596 NLEEDCFLKECGERDTMFMRINYYPPCPMPDHVLGVKPHADGSSITFLLQDKEVEGLQVL 775
Query: 234 VDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENE 293
D++W VP IPDALV+N+GDQ++IMSNGIF+SP+HRV+ N E+ R++VAMF P+ E E
Sbjct: 776 KDNQWFKVPIIPDALVINVGDQIEIMSNGIFQSPVHRVVINEEKERLTVAMFCIPDSEQE 955
Query: 294 IGPVEGLINETR 305
I PV+ L++ T+
Sbjct: 956 IKPVDKLVDGTK 991
>TC86474 weakly similar to GP|6016680|gb|AAF01507.1| putative
leucoanthocyanidin dioxygenase {Arabidopsis thaliana},
partial (20%)
Length = 1395
Score = 216 bits (550), Expect = 1e-56
Identities = 107/283 (37%), Positives = 174/283 (60%), Gaps = 2/283 (0%)
Frame = +1
Query: 37 IPIIDVSLLS--SEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEK 94
+P+ID LLS +++E KL A G FQ + HGM + ++++V FF L +EEK
Sbjct: 235 VPVIDFGLLSHGNKNELLKLDIACKEWGFFQIVNHGMEIDLMQRLKDVVAEFFDLSIEEK 414
Query: 95 QKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF 154
KYA ++ +GYG+ VVS+KQ+LDW +L L V+P R+ WPE P +++ +
Sbjct: 415 DKYAMPPDDIQGYGHTSVVSEKQILDWCDQLILLVYPTRFRKPQFWPETPEKLKDTIEAY 594
Query: 155 STKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTD 214
S+++K + + L+ +++ LEE L E + R N+YPPC+ P+ V+G+ PH+D
Sbjct: 595 SSEIKRVGEELINSLSLIFGLEEHVLLGLHKEVLQGL-RVNYYPPCNTPEQVIGLTPHSD 771
Query: 215 RSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTN 274
S +T+++QD +V GL+V WV + IP+ALVVNLGD ++++SNG +KS HR +TN
Sbjct: 772 ASTVTIVMQDDDVTGLEVRYKGNWVPINPIPNALVVNLGDVIEVLSNGKYKSVEHRAMTN 951
Query: 275 TERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNNYGD 317
+ R S F P + E+GP + +I++ P++Y+ + YG+
Sbjct: 952 KNKRRTSFVSFLFPRDDAELGPFDHMIDDQNPKMYKEI-TYGE 1077
>TC77355 weakly similar to PIR|T05552|T05552 SRG1 protein-related protein
F24A6.150 - Arabidopsis thaliana, partial (44%)
Length = 1359
Score = 215 bits (547), Expect = 2e-56
Identities = 121/322 (37%), Positives = 197/322 (60%), Gaps = 8/322 (2%)
Frame = +1
Query: 4 SVQEM-SMDSDEPPSAYV-----VERNSFGSKDSSTLIPIPIIDVSLLSSE--DEQGKLR 55
+VQEM ++ + P+ YV +E+ ++ + SS IP+ID + LS+ +E KL
Sbjct: 91 NVQEMVKVNPLQVPTKYVRNEEEMEKVNYMPQLSSE---IPVIDFTQLSNGNMEELLKLE 261
Query: 56 SALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSK 115
A G FQ + HG+ + ++R+ + FF LP+EEK+KYA N+ +GYG + VVS+
Sbjct: 262 IACKEWGFFQMVNHGVEKDLIQRMRDASNEFFELPMEEKEKYAMLPNDIQGYGQNYVVSE 441
Query: 116 KQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNL 175
+Q LDWS L L ++P R+L WP+ F E + +S++V+ + + LL ++ + L
Sbjct: 442 EQTLDWSDVLVLIIYPDRYRKLQFWPKTSHGFKEIIEAYSSEVRRVGEELLSFLSIIMGL 621
Query: 176 EEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVD 235
E+ + L++ ++ R N+YPPC+ P+ VLG+ PH+D + IT+L+QD +V GL++
Sbjct: 622 EKHA-LAELHKELIQGLRVNYYPPCNNPEQVLGLSPHSDTTTITLLIQDDDVLGLEIRNK 798
Query: 236 DKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIG 295
WV V I DALV+N+GD ++I+SNG +KS HRV+TN + R+S A F P + E+
Sbjct: 799 GNWVPVKPISDALVINVGDVIEILSNGKYKSVEHRVMTNQNKRRISYASFLFPRDDVEVE 978
Query: 296 PVEGLINETRPRLYRNVNNYGD 317
P + +I+ P++Y+ V YGD
Sbjct: 979 PFDHMIDAQNPKMYQKV-KYGD 1041
>TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin
dioxygenase-like protein {Arabidopsis thaliana}, partial
(82%)
Length = 1326
Score = 214 bits (546), Expect = 3e-56
Identities = 114/308 (37%), Positives = 178/308 (57%), Gaps = 14/308 (4%)
Frame = +2
Query: 16 PSAYVVERN------SFGSKDSSTLIPIPIIDVSLLSSEDEQ------GKLRSALSSAGC 63
PS Y+ R+ +F +++ I IP+ID+ LSSED ++ A G
Sbjct: 173 PSCYIKPRSQRPTKTTFATQNDHDHINIPVIDLEHLSSEDPVLRETVLKRVSEACREWGF 352
Query: 64 FQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSY 123
FQ + HG+S ++ +EV + FF LP+E K+++A + + EGYG+ V K +LDWS
Sbjct: 353 FQVVNHGISHELMESAKEVWREFFNLPLEVKEEFANSPSTYEGYGSRLGVKKGAILDWSD 532
Query: 124 RLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQ 183
L P R + WP PS + + E+ +V + +L M+ +L L+E ++
Sbjct: 533 YFFLHSMPPSLRNQAKWPATPSSLRKIIAEYGEEVVKLGGRMLELMSTNLGLKEDYLMNA 712
Query: 184 FGEQSSLVA--RFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNV 241
FG ++ L A R NFYP C +PDL LG+ PH+D G+T+LL D V GLQV + W+ V
Sbjct: 713 FGGENELGACLRVNFYPKCPQPDLTLGLSPHSDPGGMTILLPDDFVSGLQVRKGNDWITV 892
Query: 242 PTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLI 301
+P+A ++N+GDQ+Q++SN I+KS HRV+ N+ + R+S+AMF P+ + I P + L+
Sbjct: 893 KPVPNAFIINIGDQIQVLSNAIYKSVEHRVIVNSIKDRVSLAMFYNPKSDLLIEPAKELV 1072
Query: 302 NETRPRLY 309
+ RP LY
Sbjct: 1073TKERPALY 1096
>TC91229 similar to GP|9294689|dbj|BAB03055.1 gene_id:MHC9.10~similar to
oxidases {Arabidopsis thaliana}, partial (86%)
Length = 1198
Score = 209 bits (533), Expect = 9e-55
Identities = 109/285 (38%), Positives = 170/285 (59%), Gaps = 7/285 (2%)
Frame = +2
Query: 37 IPIIDVSLLSSEDEQ------GKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALP 90
+P+ID S S +++ KL +A G FQ I H + ++ I ++++ FF LP
Sbjct: 164 MPVIDFSKFSKGNKEEVFNELSKLSTACEEWGFFQVINHEIDLNLMENIEDMSREFFMLP 343
Query: 91 VEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGES 150
++EKQKY A +GYG V S+ Q LDW +L + P R +LWP+ P+ E+
Sbjct: 344 LDEKQKYPMAPGTVQGYGQAFVFSEDQKLDWCNMFALGIEPHYIRNSNLWPKKPARLSET 523
Query: 151 LVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVK 210
+ +S K++ + +LL+ +A L+LEE F FGE + R N+YP CSRPDLVLG+
Sbjct: 524 IELYSRKIRKLCQNLLKYIALGLSLEEDVFEKMFGEAVQAI-RMNYYPTCSRPDLVLGLS 700
Query: 211 PHTDRSGITVLLQDR-EVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMH 269
PH+D S +TVL Q + GLQ+L D+ WV V I +ALV+N+GD +++++NG +KS H
Sbjct: 701 PHSDGSALTVLQQAKGSPVGLQILKDNTWVPVEPISNALVINIGDTIEVLTNGKYKSVEH 880
Query: 270 RVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNN 314
R + + E+ R+S+ F P + E+GP+ L++E P Y+ N+
Sbjct: 881 RAVAHEEKDRLSIVTFYAPSYDLELGPMLELVDENHPCKYKRYNH 1015
>TC78110 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (53%)
Length = 1303
Score = 203 bits (517), Expect = 7e-53
Identities = 109/323 (33%), Positives = 186/323 (56%), Gaps = 12/323 (3%)
Frame = +2
Query: 31 SSTLIPIPIIDVSLLSSED------EQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAK 84
++ L+ +P+ID S L S+D E KL SA G FQ I HG+S++ ++ ++ AK
Sbjct: 191 TTPLLELPVIDFSKLFSQDLTIKGLELDKLHSACKEWGFFQLINHGVSTSLVENVKMGAK 370
Query: 85 HFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENP 144
F+ LP+EEK+K+++ + EGYG V+S++Q LDW+ + P R+ L+P+ P
Sbjct: 371 EFYNLPIEEKKKFSQKEGDVEGYGQAFVMSEEQKLDWADMFFMITLPSHMRKPHLFPKLP 550
Query: 145 SDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPD 204
F + L +S ++K + ++ MA +L ++ FGE + R N+YPPC +P+
Sbjct: 551 LPFRDDLETYSAELKKLAIQIIDFMANALKVDAKEIRELFGEGTQST-RINYYPPCPQPE 727
Query: 205 LVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIF 264
LV+G+ H+D G+T+LLQ E++GLQ+ D W+ V +P+A ++NLGD ++I++NGI+
Sbjct: 728 LVIGLNSHSDGGGLTILLQGNEMDGLQIKKDGFWIPVKPLPNAFIINLGDMLEIITNGIY 907
Query: 265 KSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYR--NVNNYGDINYRC 322
S HR N ++ R+S+A F P + P L+ P L++ V ++ Y+
Sbjct: 908 PSIEHRATVNLKKERLSIATFYSPSSAVILRPSPTLVTPKTPALFKPIGVTDF----YKG 1075
Query: 323 Y----QEGKIALETVQIAHNSDK 341
Y GK L++++I + +K
Sbjct: 1076YLGKELRGKSFLDSLRIQNEDEK 1144
>TC85652 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (50%)
Length = 1489
Score = 197 bits (500), Expect = 6e-51
Identities = 102/313 (32%), Positives = 181/313 (57%), Gaps = 4/313 (1%)
Frame = +1
Query: 30 DSSTLIPIPIIDVSLLSSED--EQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFF 87
++ +L +P+ID+ L ED E KL A G FQ I HG++++ ++K++ K F
Sbjct: 379 NTDSLPQVPVIDLGKLLGEDATELEKLDLACKEWGFFQIINHGVNTSLIEKVKIGIKEFL 558
Query: 88 ALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDF 147
+LPVEEK+K+ + N+ EG+G VVS Q L+W+ + P ++R L+P F
Sbjct: 559 SLPVEEKKKFWQTPNDMEGFGQMFVVSDDQKLEWADLFLITTLPLDERNPRLFPSIFQPF 738
Query: 148 GESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVL 207
++L + ++V+ + ++ M ++L +E F + + R+N YPPC +P+ V+
Sbjct: 739 RDNLEIYCSEVQKLCFTIISQMEKALKIESNEVTELFNHITQAM-RWNLYPPCPQPENVI 915
Query: 208 GVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSP 267
G+ PH+D +T+LLQ E+EGLQ+ D +W+ V +P+A V+N+GD ++I++NGI++S
Sbjct: 916 GLNPHSDVGALTILLQANEIEGLQIRKDGQWIPVQPLPNAFVINIGDMLEIVTNGIYRSI 1095
Query: 268 MHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRN--VNNYGDINYRCYQE 325
HR N+E+ R+S+A F+ P + P L+ RP +++ V Y + E
Sbjct: 1096EHRATVNSEKERISIAAFHRPHVNTILSPRPSLVTPERPASFKSIAVGEYLKAYFSRKLE 1275
Query: 326 GKIALETVQIAHN 338
GK L+ +++ ++
Sbjct: 1276GKSCLDVMRLEND 1314
>TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoanthocyanidin
dioxygenase {Arabidopsis thaliana}, partial (81%)
Length = 1686
Score = 197 bits (500), Expect = 6e-51
Identities = 111/306 (36%), Positives = 164/306 (53%), Gaps = 9/306 (2%)
Frame = +2
Query: 13 DEPPSAYVVERNSFGSKDSSTL--IPIPIIDVSLLSSEDEQ------GKLRSALSSAGCF 64
D P Y+ D+S+ I IPIID+ L+ +D ++ A G F
Sbjct: 320 DSIPDRYIKPPTDRPIVDTSSYDDINIPIIDLGGLNGDDLDVHASILKQISDACRDWGFF 499
Query: 65 QAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYR 124
Q + HG+S +DK RE + FF LP+E KQ+YA + EGYG+ V K +LDWS
Sbjct: 500 QIVNHGVSPDLMDKARETWRQFFHLPMEAKQQYANSPTTYEGYGSRLGVEKGAILDWSDY 679
Query: 125 LSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQF 184
L P + + WP +P E E+ ++ + L++ ++ +L LEE + F
Sbjct: 680 YFLHYLPVSVKDCNKWPASPQSCREVFDEYGKELVKLSGRLMKALSLNLGLEEKILQNAF 859
Query: 185 G-EQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPT 243
G E+ R NFYP C RP+L LG+ H+D G+T+LL D +V GLQV D W+ V
Sbjct: 860 GGEEIGACMRVNFYPKCPRPELTLGLSSHSDPGGMTMLLPDDQVAGLQVRKFDNWITVNP 1039
Query: 244 IPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINE 303
+VN+GDQ+Q++SN +KS HRV+ N+++ R+S+A F P + I P++ LI
Sbjct: 1040ARHGFIVNIGDQIQVLSNATYKSVEHRVIVNSDQERLSLAFFYNPRSDIPIEPLKQLITP 1219
Query: 304 TRPRLY 309
RP LY
Sbjct: 1220ERPALY 1237
>TC85651 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (39%)
Length = 1269
Score = 196 bits (498), Expect = 1e-50
Identities = 113/341 (33%), Positives = 187/341 (54%), Gaps = 6/341 (1%)
Frame = +3
Query: 1 MSKSVQEMSMDS-DEPPSAYVV-ERNSFGSKDSSTLIPIPIIDVSLLSSED--EQGKLRS 56
++ SVQE+ + P Y+ ++S ++++L +P+ID+S L ED E L
Sbjct: 45 LAPSVQELEKQGITKVPKQYLQPNQDSILVSNTTSLPQLPVIDLSKLLCEDAIELENLDH 224
Query: 57 ALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKK 116
A G FQ I HG++ ++ I+ + FF LP+EEK+K+ + E +G+G V ++
Sbjct: 225 ACKDWGFFQLINHGVNPLLVENIKIGVQKFFNLPIEEKKKFWQTTEEMQGFGQVYVALEE 404
Query: 117 QVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLE 176
+ L W ++ FP R L P P F ++L +S +V + L+ M+++L ++
Sbjct: 405 EKLRWGDMFFIKTFPLHTRHPHLIPCIPQPFRDNLESYSLEVNKLCVTLIEFMSKALKIK 584
Query: 177 EGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDD 236
L F E+ S R N+YPPC +PD V+G+ PH+D +T+LLQ E+EGLQ+ D
Sbjct: 585 PNELLDIF-EEGSQAMRMNYYPPCPQPDQVIGLNPHSDAGTLTILLQVNEMEGLQIKKDG 761
Query: 237 KWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGP 296
W+ + + DA VVN+GD +++ +NGI++S HR N+E+ R+SVA F+ P IGP
Sbjct: 762 IWIPIRPLSDAFVVNVGDILEVQTNGIYRSIEHRATVNSEKERISVAAFHAPHMGGYIGP 941
Query: 297 VEGLINETRPRLYRNVNNYGDIN--YRCYQEGKIALETVQI 335
L+ P L++ + +N +GK L+ V+I
Sbjct: 942 TPSLVTPQSPALFKTMPTADFLNGYIASRIKGKSYLDVVRI 1064
>TC78109 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (48%)
Length = 1314
Score = 154 bits (388), Expect(2) = 2e-50
Identities = 92/251 (36%), Positives = 144/251 (56%), Gaps = 10/251 (3%)
Frame = +3
Query: 5 VQEMSMDS-DEPPSAYVV---ERNSFGSKDSSTLIP-IPIIDVS-LLSSED----EQGKL 54
VQE++ ++ + P YV +R S +ST +P +P+ID+S LLSS D E KL
Sbjct: 81 VQELAKEALTKVPDRYVRPHHDRPILSSTTTSTPLPQLPVIDLSKLLSSHDLNEPELKKL 260
Query: 55 RSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVS 114
A G FQ I HG+S + ++ +++ A+ FF LP+EEK+K+ + + EGYG VVS
Sbjct: 261 HYACKEWGFFQLINHGVSESLMENVKKGAEEFFNLPMEEKKKFGQTEGDVEGYGQAFVVS 440
Query: 115 KKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLN 174
++Q LDW+ L L P KR+ L+ P F L + K++++ ++ MA SL
Sbjct: 441 EEQKLDWADMLVLFTLPPHKRKPHLFSNIPLPFRVDLENYCEKMRTLAIQIMDLMAHSLA 620
Query: 175 LEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLV 234
++ FG Q++ R N+YPPC +P+ V+G+ H+D G+T+LLQ E++GLQ+
Sbjct: 621 VDPMEIREFFG-QATQSTRMNYYPPCPQPEFVIGLNSHSDGGGLTILLQGNEMDGLQIKK 797
Query: 235 DDKWVNVPTIP 245
D W+ T P
Sbjct: 798 DGLWIPC*TPP 830
Score = 63.5 bits (153), Expect(2) = 2e-50
Identities = 31/104 (29%), Positives = 58/104 (54%), Gaps = 2/104 (1%)
Frame = +1
Query: 240 NVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEG 299
+V +P+A ++NLGD +++M+NGIF+S HR N+E+ R S+A F P + P
Sbjct: 814 HVKPLPNAFIINLGDMLEMMTNGIFRSIEHRATVNSEKERFSIASFYSPAFNTILSPAPS 993
Query: 300 LINETRPRLYRNVNNYGDINYRCYQE--GKIALETVQIAHNSDK 341
++ P +++ ++ QE GK L++++I +D+
Sbjct: 994 IVTPNTPAVFKRISAGEYFKGYLAQELCGKSFLDSIRIQAENDQ 1125
>TC80530 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis
thaliana, partial (52%)
Length = 1219
Score = 195 bits (496), Expect = 2e-50
Identities = 104/316 (32%), Positives = 178/316 (55%), Gaps = 4/316 (1%)
Frame = +2
Query: 1 MSKSVQEMSMDS-DEPPSAYVVERNSFGSKDSSTLIP-IPIIDVSLLSSED--EQGKLRS 56
++ SVQ+++ + P Y+ ++T P +PIID L ED E KL +
Sbjct: 20 LAPSVQKLAEQGITKVPEQYLQPNQDSILVSNTTSSPQLPIIDFDKLLCEDGIELEKLDN 199
Query: 57 ALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKK 116
A G FQ I HG++ + ++ ++ + FF LP+EEK+K+ + E +G+G V ++
Sbjct: 200 ACKEWGFFQLINHGVNPSLVESVKIGVQQFFHLPMEEKKKFWQTEEELQGFGQVYVALEE 379
Query: 117 QVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLE 176
Q L W ++ FP R L P P F + +S ++K + ++ M ++L ++
Sbjct: 380 QKLRWGDMFYVKTFPLHIRLPHLIPCMPQPFRDDFENYSLELKKLCFKIIERMTKALKIQ 559
Query: 177 EGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDD 236
+ + L F E+ R N+YPPC +PD V+G+ PH+D S +T+LLQ E++GLQ+ D
Sbjct: 560 QPNELLDFFEEGDQSIRMNYYPPCPQPDQVIGLNPHSDASALTILLQVNEMQGLQIKKDG 739
Query: 237 KWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGP 296
WV + +P+A VVN+GD ++IM+NGI++S HR N+E+ R+SVA F+ + ++GP
Sbjct: 740 MWVPINPLPNAFVVNIGDLLEIMTNGIYRSIEHRATANSEKERISVAGFHNIQMGRDLGP 919
Query: 297 VEGLINETRPRLYRNV 312
L+ P +++ +
Sbjct: 920 APSLVTPETPAMFKTI 967
>TC78010 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1321
Score = 194 bits (492), Expect = 5e-50
Identities = 109/316 (34%), Positives = 178/316 (55%), Gaps = 6/316 (1%)
Frame = +3
Query: 4 SVQEM-SMDSDEPPSAYVVERNSFGSKDSST--LIPIPIIDVS-LLSSEDEQG--KLRSA 57
SVQE+ + P Y+ + SST L +P+ID+S LLS EDE KL A
Sbjct: 111 SVQELVKQPITKVPERYLQQNQDPSLVVSSTKSLPQVPVIDLSKLLSEEDETELQKLDHA 290
Query: 58 LSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQ 117
G FQ I HG++ ++ +++ + FF LPVEEK+ ++ EG+G VVS+
Sbjct: 291 CKEWGFFQLINHGVNPLLVENFKKLVQDFFNLPVEEKKILSQKPGNIEGFGQLFVVSEDH 470
Query: 118 VLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEE 177
L+W+ + P R L+P P F ESL +S +K + ++ M+++L +++
Sbjct: 471 KLEWADLFHIITHPSYMRNPQLFPSIPQPFRESLEMYSLVLKKLCVMIIEFMSKALKIQK 650
Query: 178 GSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDK 237
L +F E+ R N+YPPC +PD V+G+ PH+D + +T+LLQ E+EGLQ+ D
Sbjct: 651 NELL-EFFEEGGQSMRMNYYPPCPQPDKVIGLNPHSDGTALTILLQLNEIEGLQIKKDGM 827
Query: 238 WVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPV 297
W+ + + +A V+N+GD ++IM+NGI++S HR N+E+ R+S+A F+ + PV
Sbjct: 828 WIPIKPLTNAFVINIGDMLEIMTNGIYRSIEHRATINSEKERISIATFHSARLNAILAPV 1007
Query: 298 EGLINETRPRLYRNVN 313
LI P ++ +++
Sbjct: 1008PSLITPKTPAVFNDIS 1055
>TC82784 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (59%)
Length = 1206
Score = 191 bits (486), Expect = 3e-49
Identities = 112/344 (32%), Positives = 195/344 (56%), Gaps = 9/344 (2%)
Frame = +3
Query: 4 SVQEMSMDS-DEPPSAYVVERNS-FGSKDSSTLIPIPIIDVSLLSSED----EQGKLRSA 57
SVQE++ + P+ Y+ ++ ++++++ IP+ID+ L S + E KL A
Sbjct: 24 SVQELAKEKISTVPARYIQSQHEELVINEANSILEIPVIDMKKLLSLEYGSLELSKLHLA 203
Query: 58 LSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQ 117
G FQ + H +SS+ L+K++ FF LP+ EK+K+ + EG+G V+S++Q
Sbjct: 204 CKDWGFFQLVNHDVSSSLLEKVKMEIMDFFNLPMSEKKKFWQTPQHMEGFGQAFVLSEEQ 383
Query: 118 VLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEE 177
LDW+ + PK R L+P+ P ++L +S ++K++ +L + +SL +EE
Sbjct: 384 KLDWADLFFMTTLPKHLRMPHLFPQLPLPLRDTLELYSQEIKNLAMVILGHIEKSLKMEE 563
Query: 178 GSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREV-EGLQVLVDD 236
F E + R N+YPPC +P+ V+G+ H+D G+T+LLQ E +GLQ+ +
Sbjct: 564 MEIRELF-EDGIQMMRTNYYPPCPQPEKVIGLTNHSDPVGLTILLQLNESRKGLQIRKNC 740
Query: 237 KWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGP 296
WV V +P+A +VN+GD ++I++NGI++S HR + N+E+ R+S+A F + +GP
Sbjct: 741 MWVPVKPLPNAFIVNIGDMLEIITNGIYRSIEHRAIVNSEKERLSIATFYSSRHGSILGP 920
Query: 297 VEGLINETRPRLYR--NVNNYGDINYRCYQEGKIALETVQIAHN 338
V+ LI E P ++ V Y + EGK ++ ++I H+
Sbjct: 921 VKSLITEQTPARFKKVGVEEYFTNLFARKLEGKSYIDVMRIEHD 1052
>TC85654 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (32%)
Length = 1307
Score = 129 bits (323), Expect(2) = 1e-47
Identities = 62/151 (41%), Positives = 99/151 (65%), Gaps = 2/151 (1%)
Frame = +1
Query: 193 RFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNL 252
R N+YPPC +P+ V+GV PH+D +T+LLQ +VEGLQ+ D +W++V +P+A V+N+
Sbjct: 688 RINYYPPCPQPEHVIGVSPHSDMGALTILLQANDVEGLQIRKDGQWISVQPLPNAFVINI 867
Query: 253 GDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNV 312
GD ++I++NGI++S HR N+++ R+S+A F+ + + IGP LI RP L++ +
Sbjct: 868 GDMLEILTNGIYRSIEHRGTVNSKKERISIATFHRLQMSSVIGPTPSLITAERPALFKTI 1047
Query: 313 NNYGDIN-YRCYQ-EGKIALETVQIAHNSDK 341
+ IN Y Q +GK L+ V+I DK
Sbjct: 1048SVADYINRYLSRQLDGKSNLDNVKIKVEMDK 1140
Score = 79.0 bits (193), Expect(2) = 1e-47
Identities = 49/185 (26%), Positives = 91/185 (48%), Gaps = 4/185 (2%)
Frame = +3
Query: 4 SVQEM-SMDSDEPPSAYVVERNSFGSKDSSTLIP-IPIIDVSLLSSED--EQGKLRSALS 59
SVQE+ + P Y+ ++T +P +P+I++ L S D E A
Sbjct: 108 SVQELVKQPITKVPEQYLQPNQDLVVVCNTTSLPKVPVINLHKLLSNDTIELENFDHACR 287
Query: 60 SAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVL 119
G FQ I HG+++ ++ +++ + FF LP++EK+KY + N+ +G+G VVS +Q L
Sbjct: 288 EWGFFQLINHGVNTLLVENMKKGVEQFFNLPIDEKKKYWQTPNDMQGFGQLFVVSDEQKL 467
Query: 120 DWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGS 179
+W + P + R L P P F + L + ++K + ++ M ++L ++
Sbjct: 468 EWQDMFYINTLPLDSRHPHLIPSIPKPFRDHLETYCLELKQLAFTIIGRMEKTLKIKTNE 647
Query: 180 FLSQF 184
+ F
Sbjct: 648 LVEFF 662
>TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1323
Score = 185 bits (470), Expect = 2e-47
Identities = 102/315 (32%), Positives = 174/315 (54%), Gaps = 6/315 (1%)
Frame = +3
Query: 4 SVQEM-SMDSDEPPSAYVVERNSFGSKDSSTLIP--IPIIDVSLLSSEDEQGKLRS---A 57
SVQE+ + P Y+ + SST+ +PIID++ L SE++ +L+ A
Sbjct: 174 SVQELVKQPITKVPERYLQQNQDPSLVVSSTISSQQVPIIDLNKLLSEEDGTELQKFDLA 353
Query: 58 LSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQ 117
G FQ I HG+ + ++ ++ + FF L EEK+ + + + EG+G VVS++
Sbjct: 354 CKEWGFFQLINHGVDLSLVENFKKHVQEFFNLSAEEKKIFGQKPGDMEGFGQMFVVSEEH 533
Query: 118 VLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEE 177
L+W+ + P R +L+P P F ++L ++ ++K + ++ MA++L ++
Sbjct: 534 KLEWADLFYIVTLPTYIRNPNLFPSIPQPFRDNLEMYAIEIKKLCVTIVEFMAKALKIQP 713
Query: 178 GSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDK 237
L F E+ S R N+YPPC +P+ V+G+ PH+D +T+LLQ E+EGLQV D
Sbjct: 714 NELLDFF-EEGSQAMRMNYYPPCPQPEQVIGLNPHSDVGALTILLQLNEIEGLQVRKDGM 890
Query: 238 WVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPV 297
W+ + + +A V+N+GD ++IM+NGI++S HR N E+ R+S+A FN + P
Sbjct: 891 WIPIKPLSNAFVINIGDMLEIMTNGIYRSIEHRATINAEKERISIATFNSGRLNAILCPA 1070
Query: 298 EGLINETRPRLYRNV 312
L+ P ++ NV
Sbjct: 1071PSLVTPESPAVFNNV 1115
>TC80257 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (50%)
Length = 1376
Score = 133 bits (335), Expect(2) = 3e-47
Identities = 66/201 (32%), Positives = 117/201 (57%), Gaps = 2/201 (0%)
Frame = +2
Query: 142 ENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCS 201
++P F E+L +S +++ + ++ MA +L + F + +V R N+YPPC
Sbjct: 590 KSPHHFRENLEVYSVELEKLAIKVIELMANALAINPKEMTELFNIGTQMV-RVNYYPPCP 766
Query: 202 RPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSN 261
+P+ V+G+K H+D G+T+LLQ +++GLQ+ D +W+ V +P+A +VN+GD ++I++N
Sbjct: 767 QPERVIGLKSHSDAGGLTILLQTSDIDGLQIRKDGQWIPVKPLPNAFIVNIGDMLEIITN 946
Query: 262 GIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNNYGDINYR 321
GI+ S HR N+E+ R+SVA F P + + P L+ + RP +R ++ N
Sbjct: 947 GIYPSIEHRATVNSEKERISVAAFYGPNMQAMLAPAPSLVTQERPAQFRRISVVDHFNGY 1126
Query: 322 CYQE--GKIALETVQIAHNSD 340
QE GK L ++I + +
Sbjct: 1127FSQELRGKSYLNEMKITKSEE 1189
Score = 72.8 bits (177), Expect(2) = 3e-47
Identities = 47/152 (30%), Positives = 81/152 (52%), Gaps = 8/152 (5%)
Frame = +1
Query: 4 SVQEMSMDS-DEPPSAYVVERNSFGSKDSSTLIP-IPIIDVSLLSSED------EQGKLR 55
SVQE++ + + P YV+ + ++T P +P+ID++ L S+D E KL
Sbjct: 151 SVQELAKEQLTKVPERYVLPDHETVVLSNTTSSPQVPVIDLAELLSQDINLKRHELEKLH 330
Query: 56 SALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSK 115
A G FQ + HG+S++ ++ +++ AK F L +EEK+ + E EG+G V+S+
Sbjct: 331 YACKEWGFFQLVNHGISTSLVEDMKKGAKTLFELSMEEKKNLWQIEGEMEGFGQSFVLSE 510
Query: 116 KQVLDWSYRLSLRVFPKEKRRLSLWPENPSDF 147
+Q L+W+ L P R+ L+ + P F
Sbjct: 511 EQKLEWADTFFLSTLPSHLRKPYLFNQIPPSF 606
>TC79368 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis
thaliana, partial (48%)
Length = 1325
Score = 180 bits (457), Expect = 6e-46
Identities = 89/277 (32%), Positives = 160/277 (57%), Gaps = 2/277 (0%)
Frame = +1
Query: 67 IGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLS 126
I HG+S + ++ ++ + F + PVEEK+K + + EG+G VVS+ Q L+W+
Sbjct: 439 INHGVSPSLVENLKIGVQEFMSRPVEEKKKLWQTPEDIEGFGQLFVVSENQKLEWADLFF 618
Query: 127 LRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGE 186
P R L+P P F ++L + ++K + ++ M ++L +E L +F +
Sbjct: 619 TTTLPSYARNPRLFPNIPQPFRDNLETYCLELKKVCITIITHMTKALKVEPKEML-EFFD 795
Query: 187 QSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPD 246
+ R N+YPPC +P+ V+G+ PH+D +T+LLQ ++EGLQ+ D +W++V +
Sbjct: 796 DLTQATRMNYYPPCPQPENVIGLNPHSDADCLTILLQANDIEGLQIRKDGQWISVKPLAG 975
Query: 247 ALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRP 306
A VVN+GD ++I++NGI++S HR N+E+ R+S+A F+ P+ IGP + L+ RP
Sbjct: 976 AFVVNIGDMLEILTNGIYRSIEHRATVNSEKARISIAAFHRPQMSKVIGPTQKLVTPERP 1155
Query: 307 RLYR--NVNNYGDINYRCYQEGKIALETVQIAHNSDK 341
L++ V +Y + +GK ++ ++I + + K
Sbjct: 1156ALFKTITVEDYYKAFFSRKLQGKSCIDLMRIQNENSK 1266
>CA918846 weakly similar to GP|21537316|gb ethylene-forming-enzyme-like
dioxygenase-like {Arabidopsis thaliana}, partial (44%)
Length = 721
Score = 178 bits (451), Expect = 3e-45
Identities = 87/185 (47%), Positives = 124/185 (67%)
Frame = -3
Query: 158 VKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSG 217
V + + + M + LNL+E FL + GE+ ++ R N+YPPC D LG+KPH+D S
Sbjct: 671 VWQLYEEISGPMPKLLNLKEDCFLKECGERGTMSMRTNYYPPCPMADHALGLKPHSDSSS 492
Query: 218 ITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTER 277
IT LLQD +VEGLQVL D++W VP I DA+V+N+GD M+IMSNGIF+SP+HR + N E+
Sbjct: 491 ITFLLQDNKVEGLQVLKDNQWFKVPIIHDAIVINVGDLMEIMSNGIFQSPIHRAVVNEEK 312
Query: 278 LRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQIAH 337
R+SVAM P+ E EI P++ L+NE+RP YR V +Y + YQ+GK ++ ++
Sbjct: 311 ARLSVAMLCRPDSEKEIQPIDKLVNESRPLSYRPVKDYCTKYLQYYQQGKRPIDAFKL*S 132
Query: 338 NSDKK 342
N+ K
Sbjct: 131 NAS*K 117
>TC91108 similar to PIR|T05552|T05552 SRG1 protein-related protein F24A6.150
- Arabidopsis thaliana, partial (14%)
Length = 1133
Score = 176 bits (446), Expect = 1e-44
Identities = 97/283 (34%), Positives = 160/283 (56%), Gaps = 2/283 (0%)
Frame = +2
Query: 37 IPIIDVSLLSS--EDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEK 94
+PIID LLS+ ++E KL A G FQ + HGM + ++++ F L ++EK
Sbjct: 269 LPIIDFGLLSNGNKEELLKLDFACKEWGYFQIVNHGMQIDLMQRVKDAVAEFSELSIKEK 448
Query: 95 QKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF 154
KYA ++ +GYG+ V DWS L L V+P R+ WP+ D ++ +
Sbjct: 449 IKYAMPPDDMQGYGHTSV-------DWSDYLVLLVYPTRFRKPQFWPKELKD---TIDAY 598
Query: 155 STKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTD 214
S ++K + + ++ +++ L LEE L E R N+YPPC+ P+ VLGV PH+D
Sbjct: 599 SNEIKRIGEEVINSLSLLLGLEEHGLLDLHREVHQGF-RVNYYPPCNTPEKVLGVSPHSD 775
Query: 215 RSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTN 274
I +++QD +V G ++ WV + IP ALVVN+GD ++I+SNG +KS HR +TN
Sbjct: 776 PRTIAIVMQDDDVSGTEIRYKRNWVPITPIPGALVVNVGDVIEILSNGKYKSVEHRAITN 955
Query: 275 TERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNNYGD 317
+ R + + P+ + EIG ++ +I++ P++Y++ YG+
Sbjct: 956 KNKKRTTFISYLFPQGDAEIGALDHMIDDQNPKMYKD-TTYGE 1081
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,613,533
Number of Sequences: 36976
Number of extensions: 121912
Number of successful extensions: 764
Number of sequences better than 10.0: 156
Number of HSP's better than 10.0 without gapping: 668
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 673
length of query: 342
length of database: 9,014,727
effective HSP length: 97
effective length of query: 245
effective length of database: 5,428,055
effective search space: 1329873475
effective search space used: 1329873475
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0219.11


