

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
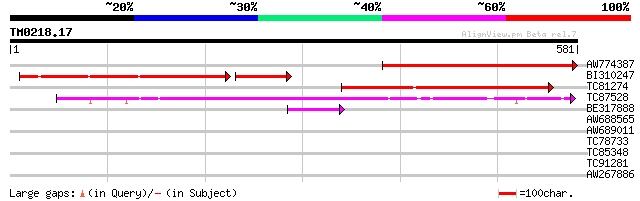
Query= TM0218.17
(581 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW774387 weakly similar to PIR|T05210|T05 hypothetical protein F... 280 8e-76
BI310247 weakly similar to PIR|T05213|T05 hypothetical protein F... 195 4e-62
TC81274 similar to GP|23429644|gb|AAN10199.1 endoxylanase {Caric... 209 2e-54
TC87528 similar to PIR|D96617|D96617 probable xylan endohydrolas... 203 1e-52
BE317888 similar to PIR|T05693|T056 hypothetical protein F20M13.... 60 2e-09
AW688565 similar to PIR|D96617|D96 probable xylan endohydrolase ... 35 0.10
AW689011 similar to PIR|D96617|D96 probable xylan endohydrolase ... 32 0.52
TC78733 similar to GP|21592813|gb|AAM64762.1 ids4-like protein {... 31 1.5
TC85348 similar to PIR|F96792|F96792 unknown protein F14G6.8 [im... 30 2.6
TC91281 similar to PIR|E71427|E71427 hypothetical protein - Arab... 30 2.6
AW267886 28 9.8
>AW774387 weakly similar to PIR|T05210|T05 hypothetical protein F17I5.10 -
Arabidopsis thaliana (fragment), partial (19%)
Length = 658
Score = 280 bits (717), Expect = 8e-76
Identities = 135/199 (67%), Positives = 156/199 (77%)
Frame = -2
Query: 383 FLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMRAG 442
F+NE+NTIEYSGD SP NY+KK+++I+Q + I AIGLQ HFASGQPNLAYMR+G
Sbjct: 657 FMNEYNTIEYSGDKVVSPPNYLKKLEEIMQSGEATEILFAIGLQGHFASGQPNLAYMRSG 478
Query: 443 LDLLGATGFPIWLTETSIDPQPHQEDYFDWILREGYAHPAVQGIIMFMGPVQAGFKKAPL 502
LD LG GFPIWLTE S+DPQP+Q +YF+ +LRE Y+HPAV+GIIMF GP QAGF L
Sbjct: 477 LDFLGNIGFPIWLTEASLDPQPNQAEYFEEVLREAYSHPAVEGIIMFAGPAQAGFNSTLL 298
Query: 503 ADENFKNTPIGDVVDMLIREWGTGPHEAKADSRGIVDISLHHGDYDVIVTHPQKQISKTL 562
AD NF+ TP G VVD LI EWG+GP+ A ADSRGIVDISLHHGDYDV THP Q SK L
Sbjct: 297 ADTNFQTTPTGQVVDNLILEWGSGPYTAIADSRGIVDISLHHGDYDVTFTHPLTQNSKKL 118
Query: 563 NLSVRKGSPQQTIQVKMHA 581
N+SVRKG Q+ I VKMHA
Sbjct: 117 NISVRKGFSQENIHVKMHA 61
>BI310247 weakly similar to PIR|T05213|T05 hypothetical protein F17I5.40 -
Arabidopsis thaliana, partial (39%)
Length = 841
Score = 195 bits (496), Expect(2) = 4e-62
Identities = 101/216 (46%), Positives = 137/216 (62%)
Frame = +1
Query: 11 LFTTSFLILISGLTVHSLSHSYDYSATTECLAEPHREHYGGGIIVNPGFDHNLEGWTVFG 70
+F +IL++G +LS YD+SA+ ECLA P + Y GGII NP + L+GWT FG
Sbjct: 10 IFLLLCVILLAGFEAEALS--YDHSASIECLAHPQKPQYNGGIIKNPELNDGLQGWTAFG 183
Query: 71 NGAIEERRSNEGNAFIVARNRTQPLDSFSQKIQLKKGMLYTFSAWLQLSEGSDTVSVVFK 130
N IE R S GN ++VA NR QP DS +QKI L+ G+ Y+ SAW+Q+SE + V+ + K
Sbjct: 184 NARIEHRESL-GNKYVVANNRNQPHDSVAQKIYLQNGLHYSLSAWIQVSEANVLVTAMVK 360
Query: 131 INGRELIRGGHVIAKHGCWTLMKGGLVANFTSPAEILFESKNPTVEIWADSVSLQPFTKE 190
GG + A+ CW+++KGGL A+ T AE+ FES N +VEIW D++SLQPFTK+
Sbjct: 361 TT-EGFKFGGAIYAEPNCWSMLKGGLTADATEVAELYFESNNTSVEIWIDNISLQPFTKK 537
Query: 191 QWRSHQEESIERVRKSKVRFQVTHINETALEGATIT 226
QW SHQE++IE+ RK V Q L A I+
Sbjct: 538 QWTSHQEQNIEKTRKKNVVVQALDEQGRPLPNARIS 645
Score = 61.6 bits (148), Expect(2) = 4e-62
Identities = 27/57 (47%), Positives = 36/57 (62%)
Frame = +3
Query: 232 LDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMM 288
L F FG +N IL+N+ Y WF SRF T F N+MKWY+ E +G++NY D M+
Sbjct: 663 LVFHFGSAMNKYILSNTAYHYWFASRFTGTAF*NEMKWYTNEYTQGRDNYFEADVML 833
>TC81274 similar to GP|23429644|gb|AAN10199.1 endoxylanase {Carica papaya},
partial (33%)
Length = 912
Score = 209 bits (533), Expect = 2e-54
Identities = 99/217 (45%), Positives = 141/217 (64%)
Frame = +2
Query: 341 ELIAWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASP 400
+LI WDV+NENLH +FFE LG+N ++ ++ H++D NT +F+N++NTIE S D ++P
Sbjct: 2 QLIGWDVINENLHSSFFESKLGQNFTSRMFNEVHNVDGNTTLFMNDYNTIEDSRDGLSTP 181
Query: 401 TNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETSI 460
YI+KI++I + + +PL IGL+ HF + PNL YMRA LD+L ATG PIW+TE +
Sbjct: 182 PKYIQKIREIQSW--NKQLPLGIGLESHFPNFPPNLPYMRASLDILAATGLPIWITELDV 355
Query: 461 DPQPHQEDYFDWILREGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNTPIGDVVDMLI 520
QP+Q Y + +LRE ++HP ++GI+M+ G + L D NFKN P GDVVD LI
Sbjct: 356 ASQPNQAGYLEQVLREAHSHPGIRGIVMWTAWSPQGCFRMCLTDNNFKNLPTGDVVDKLI 535
Query: 521 REWGTGPHEAKADSRGIVDISLHHGDYDVIVTHPQKQ 557
EWG D G + SL HGDY + ++H K+
Sbjct: 536 YEWGRKILSGTTDKNGFLKASLFHGDYKMEISHSAKK 646
>TC87528 similar to PIR|D96617|D96617 probable xylan endohydrolase isoenzyme
F9K23.10 [imported] - Arabidopsis thaliana, partial
(67%)
Length = 2080
Score = 203 bits (517), Expect = 1e-52
Identities = 152/564 (26%), Positives = 261/564 (45%), Gaps = 33/564 (5%)
Frame = +1
Query: 49 YGGGIIVNPGFDHNLEGWTVFGNGAIEERRSNE-----------------GNAFIVARNR 91
+G II N + +GW GN + + + +I+ NR
Sbjct: 283 FGVNIIENSNLSDDTKGWFTLGNCPLTVKTGSPHILPPMARESLGPHGILSGRYILVTNR 462
Query: 92 TQPLDSFSQKI--QLKKGMLYTFSAWLQL---SEGSDTVSVVFKINGRELIRGGHVIAKH 146
TQ +Q I +LK + Y SAW+++ S G V+V + + I GG
Sbjct: 463 TQTWMGPAQVITEKLKLFLTYQVSAWVRIGSSSNGPQNVNVALGADN-QWINGGQTEVSD 639
Query: 147 GCWTLMKGGLVANFTSPAEILFESKNPT--VEIWADSVSLQPFTKEQWRSHQEESIERVR 204
W + GG P +I+ + P V++ + + P + + + +++R
Sbjct: 640 DRWHEI-GGSFRIEKQPTKIMVYIQGPASGVDLMVAGLQIFPVDRHARFRYLKMQTDKIR 816
Query: 205 KSKVRFQVTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFT 264
K V + ++ ++ + +QT+ +FP G I+ + + N ++ + + F + F
Sbjct: 817 KRDVVLKFAGLDSSSYLNTMVQVRQTQNNFPIGTCISRSNIDNEDFVNFLVKHFNWAVFA 996
Query: 265 NQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEELR 324
N++KWY TE +G NY D +++L + I RGH IFW+ QWVKSL+ +L
Sbjct: 997 NELKWYWTEPQQGNLNYKDADDLLTLCQKYKIQTRGHCIFWEVDGTVQQWVKSLNKNDLM 1176
Query: 325 KAAAKRINSVASRYSGELIAWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFL 384
A R+ S+ +RY G+ +DV NE LH +F++ LG++ A + TA+ LD + +F+
Sbjct: 1177TAVQNRLTSLLTRYKGKFSHYDVNNEMLHGSFYQDRLGKDIRANMFKTANQLDLSATLFV 1356
Query: 385 NEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMRAGLD 444
N+++ IE D + P YI+ I + + G IG+Q H S P + + LD
Sbjct: 1357NDYH-IEDGCDTRSCPNKYIEHILDLQEQGAPVG---GIGIQGHIDS--PVGPVVCSSLD 1518
Query: 445 LLGATGFPIWLTETSIDPQPH--QEDYFDWILREGYAHPAVQGIIMFMGPVQAGFKKAPL 502
LG G PIW TE + + D + +LRE AHPAV+GI+++ GF + +
Sbjct: 1519KLGILGLPIWFTELDVSSMNEYVRGDDLEVMLREAMAHPAVEGIMLW------GFWELFM 1680
Query: 503 ADENFKNTPI-GDVVD------MLIREWGTGPHEAKADSRGIVDISLHHGDYDVIVTHPQ 555
+ +N GD+ + L +EW + H + +G + +G Y+V + P
Sbjct: 1681SRDNAHLVNAEGDINEAGKRFLALKQEWLSHSH-GHVNEQGQFNFRGFYGTYNVEIVTPS 1857
Query: 556 KQISKTLNLSVRKGSPQQTIQVKM 579
K+ISKT L KG + + +
Sbjct: 1858KKISKTFVLD--KGDSPMEVSIDL 1923
>BE317888 similar to PIR|T05693|T056 hypothetical protein F20M13.210 -
Arabidopsis thaliana, partial (13%)
Length = 205
Score = 60.1 bits (144), Expect = 2e-09
Identities = 26/59 (44%), Positives = 35/59 (59%)
Frame = +1
Query: 285 DAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELI 343
D MM + N I RGHNIFW+DPK P WV +L+ +L+ A RI + ++Y E I
Sbjct: 25 DQMMQFVRANKIIARGHNIFWEDPKYNPAWVLNLTGTQLKSAVNSRIQRLMNQYKAEFI 201
>AW688565 similar to PIR|D96617|D96 probable xylan endohydrolase isoenzyme
F9K23.10 [imported] - Arabidopsis thaliana, partial
(11%)
Length = 614
Score = 34.7 bits (78), Expect = 0.10
Identities = 35/141 (24%), Positives = 64/141 (44%), Gaps = 15/141 (10%)
Frame = +2
Query: 3 VSAEGCSLLFTTSFLILISGLTVHS----LSHSYDYSATTECLAEPHREHYGGGIIVNPG 58
+++ C + F+ F I S +H+ +S + + T+C++ II+NP
Sbjct: 68 ITSSTCYIHFSLCFKI--SSKFLHNNF*CISPLFQKTTNTKCVSTDDEN-----IILNPE 226
Query: 59 FDHNLEGWTVFGNG---AIEERRSN------EGNAFIVARNRTQPLDSFSQKI--QLKKG 107
F+ L WT G G A+ E +N G F A RTQ + Q+I ++++
Sbjct: 227 FEDGLNNWT--GRGCKIAVHESMANGKILPKSGKFFASATERTQSWNGIQQEITGRVQRK 400
Query: 108 MLYTFSAWLQLSEGSDTVSVV 128
+ Y +A +++ + T S V
Sbjct: 401 LAYEVTALVRIFGNNVTTSDV 463
>AW689011 similar to PIR|D96617|D96 probable xylan endohydrolase isoenzyme
F9K23.10 [imported] - Arabidopsis thaliana, partial
(12%)
Length = 591
Score = 32.3 bits (72), Expect = 0.52
Identities = 29/105 (27%), Positives = 46/105 (43%), Gaps = 11/105 (10%)
Frame = +2
Query: 53 IIVNPGFDHNLEGWTVFGNG---AIEERRSN------EGNAFIVARNRTQPLDSFSQKI- 102
II+NP F+ L WT G G A+ E +N G F A RTQ + Q+I
Sbjct: 203 IILNPEFEDGLNNWT--GRGCKIAVHESMANGKILPKSGKFFASATERTQSWNGIQQEIT 376
Query: 103 -QLKKGMLYTFSAWLQLSEGSDTVSVVFKINGRELIRGGHVIAKH 146
++++ + Y +A ++ + T S V + R I +H
Sbjct: 377 GRVQRKLAYEVTALXRIFGNNVTTSDVRATLYVQKTRSSRAIYRH 511
>TC78733 similar to GP|21592813|gb|AAM64762.1 ids4-like protein {Arabidopsis
thaliana}, partial (83%)
Length = 1260
Score = 30.8 bits (68), Expect = 1.5
Identities = 27/123 (21%), Positives = 53/123 (42%)
Frame = -1
Query: 117 QLSEGSDTVSVVFKINGRELIRGGHVIAKHGCWTLMKGGLVANFTSPAEILFESKNPTVE 176
Q +EG+ ++ ++F+ + G VI + ++ +ANF I+ NP ++
Sbjct: 687 QANEGTSSLVILFQYFH*PCVIQGTVILQQDHLSMEIHNFLANFHHLFAIILHFSNPILQ 508
Query: 177 IWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQVTHINETALEGATITTKQTKLDFPF 236
+ + L F E+ E +++V S+V F + H + + G T +
Sbjct: 507 LLESNNVLFFFLNEEVIEFLEFILQQV--SEVTFLLRHFSGVGVAGINFKTFHRTVIGGG 334
Query: 237 GCG 239
GCG
Sbjct: 333 GCG 325
>TC85348 similar to PIR|F96792|F96792 unknown protein F14G6.8 [imported] -
Arabidopsis thaliana, partial (36%)
Length = 878
Score = 30.0 bits (66), Expect = 2.6
Identities = 15/46 (32%), Positives = 20/46 (42%)
Frame = +1
Query: 21 SGLTVHSLSHSYDYSATTECLAEPHREHYGGGIIVNPGFDHNLEGW 66
S L++H + YDYSA L PH ++ G N E W
Sbjct: 454 SSLSIHFIPFQYDYSA----LHSPHHHERSNPLLWKYGRGQNFESW 579
>TC91281 similar to PIR|E71427|E71427 hypothetical protein - Arabidopsis
thaliana, partial (36%)
Length = 1107
Score = 30.0 bits (66), Expect = 2.6
Identities = 19/60 (31%), Positives = 30/60 (49%), Gaps = 4/60 (6%)
Frame = +3
Query: 216 NETALEGATITTKQTKLDFPFG--CGINNNILTNSEYQKWFLSRFKYTTF--TNQMKWYS 271
NE A E A++ Q P G CG++ N NS+Y+K + +R + TN W++
Sbjct: 471 NEFAHEWASLKNLQVPNPTPCGDNCGVSINWHVNSDYEKGWTARITIFNWGETNYADWFA 650
>AW267886
Length = 590
Score = 28.1 bits (61), Expect = 9.8
Identities = 19/45 (42%), Positives = 25/45 (55%), Gaps = 2/45 (4%)
Frame = +3
Query: 294 NGISVRGHNIFWDDPKQQPQW-VKSLS-PEELRKAAAKRINSVAS 336
NGISV HN +WDD QP + SLS LR ++ S++S
Sbjct: 267 NGISV--HNTYWDDTLAQPIFPTTSLSISSSLRWTETTKVKSLSS 395
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,401,065
Number of Sequences: 36976
Number of extensions: 250977
Number of successful extensions: 1196
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 1180
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1191
length of query: 581
length of database: 9,014,727
effective HSP length: 101
effective length of query: 480
effective length of database: 5,280,151
effective search space: 2534472480
effective search space used: 2534472480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0218.17


