

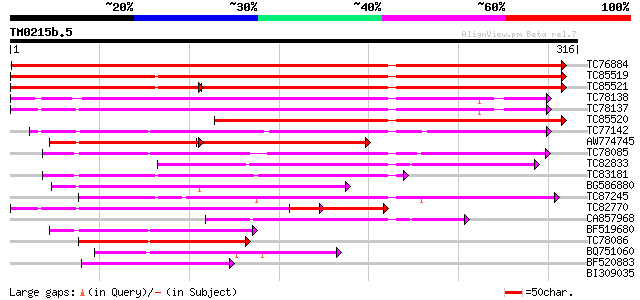
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.5
(316 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76884 similar to SP|P26690|6DCS_SOYBN NAD(P)H dependent 6'-deo... 386 e-108
TC85519 homologue to GP|537298|gb|AAB41556.1|| chalcone reductas... 290 5e-79
TC85521 homologue to GP|537298|gb|AAB41556.1|| chalcone reductas... 199 4e-74
TC78138 similar to GP|2792155|emb|CAA11226.1 chalcone reductase ... 195 2e-50
TC78137 similar to GP|2792155|emb|CAA11226.1 chalcone reductase ... 192 1e-49
TC85520 homologue to GP|537298|gb|AAB41556.1|| chalcone reductas... 184 5e-47
TC77142 homologue to GP|9927482|emb|CAC04887.1 unnamed protein p... 151 4e-37
AW774745 weakly similar to GP|2792155|emb chalcone reductase {Se... 84 4e-32
TC78085 similar to GP|9927482|emb|CAC04887.1 unnamed protein pro... 128 3e-30
TC82833 similar to PIR|T48188|T48188 aldose reductase-like prote... 127 6e-30
TC83181 similar to PIR|T02543|T02543 aldehyde dehydrogenase homo... 120 9e-28
BG586880 weakly similar to GP|2792155|emb chalcone reductase {Se... 117 4e-27
TC87245 similar to GP|10334991|gb|AAG15839.2 NADPH-dependent man... 106 1e-23
TC82770 homologue to GP|2792155|emb|CAA11226.1 chalcone reductas... 103 1e-22
CA857968 similar to PIR|T48188|T48 aldose reductase-like protein... 94 9e-20
BF519680 weakly similar to GP|13160399|em aldose reductase {Digi... 84 5e-17
TC78086 similar to GP|9927482|emb|CAC04887.1 unnamed protein pro... 72 4e-13
BQ751060 similar to GP|14330324|emb aldo-keto reductase {Gallus ... 61 5e-10
BF520883 weakly similar to PIR|S78113|S78 aldehyde reductase (NA... 60 8e-10
BI309035 weakly similar to PIR|C82294|C822 oxidoreductase Tas a... 40 0.002
>TC76884 similar to SP|P26690|6DCS_SOYBN NAD(P)H dependent 6'-deoxychalcone
synthase (EC 1.1.-.-). [Soybean] {Glycine max}, partial
(95%)
Length = 1256
Score = 386 bits (991), Expect = e-108
Identities = 207/311 (66%), Positives = 243/311 (77%), Gaps = 2/311 (0%)
Frame = +2
Query: 2 ATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVE 61
AT TVP+VVL SSTG +++ VMGL TAPEA + ++T D VLEA+KQG++HFDAAAAY VE
Sbjct: 77 ATPTVPEVVLPSSTGQRKMPVMGLGTAPEATSKVTTKDAVLEAIKQGYRHFDAAAAYGVE 256
Query: 62 QSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIH 121
+SVG AIAE + LI+S+D LFVTSK+WV DNHPELIVPALQK LRTLQLE LDL LIH
Sbjct: 257 KSVGEAIAEALKLGLISSRDELFVTSKLWVTDNHPELIVPALQKSLRTLQLENLDLILIH 436
Query: 122 WPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFA 181
WP TK G VKYPIEV EI+EF M+GV +S+EE Q+LGLTKAIG +N SIKKL+KL FA
Sbjct: 437 WPITTKPGEVKYPIEVSEIVEFDMKGVWTSLEECQKLGLTKAIGASNFSIKKLEKLLSFA 616
Query: 182 TISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPLRMGVSRGANLAMDNDIQKE 241
TI P ++QVEV+LGWQQ KLR FCKEK + + AFSPLR G SRGANL MDNDI KE
Sbjct: 617 TIPPAVNQVEVNLGWQQEKLRAFCKEKGIV----VTAFSPLRKGASRGANLVMDNDILKE 784
Query: 242 L-DTHGKTIAQICL*WIYKH-VNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQER 299
L D HGKTIAQICL W+Y+ + F + + L IF SLTE+D++KISEIHQER
Sbjct: 785 LADAHGKTIAQICLRWLYEQGLTFVVKSYDKERMNQNLQIFDWSLTEDDYKKISEIHQER 964
Query: 300 LIKEPTKPLLN 310
LIK PTKPLL+
Sbjct: 965 LIKGPTKPLLD 997
>TC85519 homologue to GP|537298|gb|AAB41556.1|| chalcone reductase {Medicago
sativa subsp. sativa}, complete
Length = 1167
Score = 290 bits (742), Expect = 5e-79
Identities = 151/312 (48%), Positives = 212/312 (67%), Gaps = 2/312 (0%)
Frame = +1
Query: 1 MATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAV 60
M + +P VL +++ ++ V+G+ +AP+ T D ++EA+KQG++HFD AAAY
Sbjct: 76 MGSVEIPTKVLTNTSSQLKMPVVGMGSAPDFTCKKDTKDAIIEAIKQGYRHFDTAAAYGS 255
Query: 61 EQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLI 120
EQ++G A+ E + L+ +D LFVTSK+WV +NHP L++PALQK L+TLQL+YLDL+LI
Sbjct: 256 EQALGEALKEAIELGLVTRQD-LFVTSKLWVTENHPHLVIPALQKSLKTLQLDYLDLYLI 432
Query: 121 HWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCF 180
HWP +++ G +PI+V +++ F ++GV SMEE +LGLTKAIGV+N S+KKL+ L
Sbjct: 433 HWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVSNFSVKKLENLLSV 612
Query: 181 ATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPLRMGVSRGANLAMDNDIQK 240
ATI P ++QVE++L WQQ KLR+FC + + AFSPLR G SRG N M+ND+ K
Sbjct: 613 ATILPAVNQVEMNLAWQQKKLREFCNANGIV----LTAFSPLRKGASRGPNEVMENDMLK 780
Query: 241 EL-DTHGKTIAQICL*WIYKH-VNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQE 298
E+ D HGK++AQI L W+Y+ V F + + L IF SLT+ D EKI +I Q
Sbjct: 781 EIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQN 960
Query: 299 RLIKEPTKPLLN 310
RLI PTKP LN
Sbjct: 961 RLIPGPTKPGLN 996
>TC85521 homologue to GP|537298|gb|AAB41556.1|| chalcone reductase {Medicago
sativa subsp. sativa}, complete
Length = 1508
Score = 199 bits (505), Expect(2) = 4e-74
Identities = 104/205 (50%), Positives = 139/205 (67%), Gaps = 2/205 (0%)
Frame = +3
Query: 108 RTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVN 167
RTLQL+YLDL+LIHWP +++ G +PI+V +++ F ++GV SMEE +LGLTKAIGV+
Sbjct: 633 RTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVS 812
Query: 168 NLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPLRMGVS 227
N S+KKL+ L ATI P ++QVE++L WQQ KLR+FC + + AFSPLR G S
Sbjct: 813 NFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIV----LTAFSPLRKGAS 980
Query: 228 RGANLAMDNDIQKEL-DTHGKTIAQICL*WIYKH-VNFCDEEL*QGEDATKLAIFGCSLT 285
RG N M+ND+ KE+ D HGK++AQI L W+Y+ V F + + L IF SLT
Sbjct: 981 RGPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLT 1160
Query: 286 ENDFEKISEIHQERLIKEPTKPLLN 310
+ D EKI +I Q RLI PTKP +N
Sbjct: 1161KEDHEKIDQIKQNRLIPGPTKPGIN 1235
Score = 97.1 bits (240), Expect(2) = 4e-74
Identities = 47/108 (43%), Positives = 74/108 (68%)
Frame = +2
Query: 1 MATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAV 60
M + +P VL +++ ++ V+G+ +AP+ T D ++EA+KQG++HFD AAAY
Sbjct: 212 MGSVEIPTKVLTNTSSQLKMPVVGMGSAPDFTCKKDTKDAIIEAIKQGYRHFDTAAAYGS 391
Query: 61 EQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLR 108
EQ++G A+ E + L+ +D LFVTSK+WV +NHP L++PALQK L+
Sbjct: 392 EQALGEALKEAIELGLVTRQD-LFVTSKLWVTENHPHLVIPALQKSLK 532
>TC78138 similar to GP|2792155|emb|CAA11226.1 chalcone reductase {Sesbania
rostrata}, partial (83%)
Length = 1289
Score = 195 bits (496), Expect = 2e-50
Identities = 117/310 (37%), Positives = 182/310 (57%), Gaps = 8/310 (2%)
Frame = +2
Query: 1 MATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAV 60
M T VP+VVL S +++ ++G T I +L+A+ G++HFD AA Y
Sbjct: 53 METKKVPEVVLNSG---KKMPMIGFGTGTTPPQQI-----MLDAIDIGYRHFDTAALYGT 208
Query: 61 EQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLI 120
E+ +G A+++ + L+ ++D LF+TSK+W D +L++PAL+ L+ L LEY+DL+LI
Sbjct: 209 EEPLGQAVSKALELGLVKNRDELFITSKLWCTDAQHDLVLPALKTTLKNLGLEYVDLYLI 388
Query: 121 HWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCF 180
HWP KQ A + +++ F ++G +MEE RLGL K+IGV+N +KKL L
Sbjct: 389 HWPVRLKQDAESLKFKKEDMIPFDIKGTWEAMEECYRLGLAKSIGVSNFGVKKLSILLEN 568
Query: 181 ATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDNDIQ 239
A I+P ++QVE++ WQQ KLR+FCK+K ++ + A+SPL +S G+ M+N I
Sbjct: 569 AEIAPAVNQVEMNPSWQQGKLREFCKQKGIH----VSAWSPLGGYKLSWGSPTVMENPIL 736
Query: 240 KEL-DTHGKTIAQICL*WIYKH------VNFCDEEL*QGEDATKLAIFGCSLTENDFEKI 292
E+ + K++AQI L WIY+ +F E + Q + IF L + + +KI
Sbjct: 737 HEIAEARKKSVAQIALRWIYQQGAIPIVKSFNKERMKQ-----NIEIFDWELNQEELDKI 901
Query: 293 SEIHQERLIK 302
S+IHQ R K
Sbjct: 902 SQIHQSRFQK 931
>TC78137 similar to GP|2792155|emb|CAA11226.1 chalcone reductase {Sesbania
rostrata}, partial (93%)
Length = 1029
Score = 192 bits (488), Expect = 1e-49
Identities = 113/310 (36%), Positives = 186/310 (59%), Gaps = 8/310 (2%)
Frame = +3
Query: 1 MATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAV 60
M VP+V+L S ++ +G ST+P + + T ++++A+K G++HFD A+ Y
Sbjct: 12 MEINKVPEVILNSGKN-MPMIGLGTSTSPSPPHEVLT-SILVDAIKIGYRHFDTASIYNT 185
Query: 61 EQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLI 120
E+ +G A+++ + L+ ++D LFVTSK+W D H +L++PAL+ L+ L L Y+DL+LI
Sbjct: 186 EEPLGQAVSKALELGLVKNRDELFVTSKLWCTDAHHDLVLPALKSTLKNLGLGYVDLYLI 365
Query: 121 HWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCF 180
HWP KQ + + + + F ++G SME+ RLG+ K+IGV+N IKKL L
Sbjct: 366 HWPVRLKQDVEGHNFKGEDTIPFDIKGTWESMEDCYRLGIAKSIGVSNFGIKKLSMLLEN 545
Query: 181 ATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDNDIQ 239
A I+P ++QVE++ W Q KLR+FCK+K ++ + A+SPL +S G+ M+N I
Sbjct: 546 AEIAPAVNQVEMNSSWHQGKLREFCKQKGIH----VSAWSPLGGYKLSWGSPAVMENLIL 713
Query: 240 KEL-DTHGKTIAQICL*WIYKH------VNFCDEEL*QGEDATKLAIFGCSLTENDFEKI 292
+++ + K++AQI L WIY+ +F E + Q + IF L + + +KI
Sbjct: 714 RKIAEARKKSVAQIALRWIYQQGVIPIVKSFNKERMKQ-----NIEIFDWELNQEELDKI 878
Query: 293 SEIHQERLIK 302
++I Q RL+K
Sbjct: 879 NQIPQCRLLK 908
>TC85520 homologue to GP|537298|gb|AAB41556.1|| chalcone reductase {Medicago
sativa subsp. sativa}, partial (63%)
Length = 780
Score = 184 bits (466), Expect = 5e-47
Identities = 97/198 (48%), Positives = 131/198 (65%), Gaps = 2/198 (1%)
Frame = +1
Query: 115 LDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKL 174
L L+LIHWP +++ G +PI+V +++ F ++GV SMEE +LGLTKAIGV+N S+KKL
Sbjct: 13 LGLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVSNFSVKKL 192
Query: 175 DKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPLRMGVSRGANLAM 234
+ L ATI P ++QVE++L WQQ KLR+FC + + AFSPLR G SRG N M
Sbjct: 193 ENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIV----LTAFSPLRKGASRGPNEVM 360
Query: 235 DNDIQKEL-DTHGKTIAQICL*WIYKH-VNFCDEEL*QGEDATKLAIFGCSLTENDFEKI 292
+ND+ KE+ D HGK++AQI L W+Y+ V F + + L IF SLT+ D EKI
Sbjct: 361 ENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEKI 540
Query: 293 SEIHQERLIKEPTKPLLN 310
+I Q RLI PTKP +N
Sbjct: 541 DQIKQNRLIPGPTKPGIN 594
>TC77142 homologue to GP|9927482|emb|CAC04887.1 unnamed protein product
{Medicago sativa}, complete
Length = 1350
Score = 151 bits (381), Expect = 4e-37
Identities = 101/294 (34%), Positives = 162/294 (54%), Gaps = 3/294 (1%)
Frame = +3
Query: 12 QSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEE 71
Q +TG +I +GL T +++ V AV+ G++H D A AY + +G+A+ +
Sbjct: 39 QLNTG-AKIPSVGLGTWQAEPGVVAK--AVTTAVQVGYRHIDCAEAYKNQSEIGSALKKL 209
Query: 72 HQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAV 131
+ +I ++ L++TSK+W D+HPE + AL K L LQL+YLDL+LIHWP + K+G
Sbjct: 210 FEDGVI-KREELWITSKLWCSDHHPEDVPKALDKTLNDLQLDYLDLYLIHWPVSMKRGTG 386
Query: 132 KYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVE 191
++ E L+ + + ++E G KAIGV+N S KKL L A + P ++QVE
Sbjct: 387 EFKAENLDRAD--IPSTWKALEALYDSGKAKAIGVSNFSTKKLQDLLDVARVPPAVNQVE 560
Query: 192 VDLGWQQVKLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDNDIQKEL-DTHGKTI 249
+ GWQQ KL FC+ K ++ + +SPL GV + + N + KE+ + GKT
Sbjct: 561 LHPGWQQAKLHAFCESKGIH----VSGYSPLGSPGVLKSD--ILKNPVVKEIAEKLGKTP 722
Query: 250 AQICL*W-IYKHVNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQERLIK 302
Q+ L W + + + + L ++ S+ E+ F K SEI+Q++LIK
Sbjct: 723 GQVALRWGLQAGHSVLPKSTNEARIKENLDVYDWSIPEDLFPKFSEINQDKLIK 884
>AW774745 weakly similar to GP|2792155|emb chalcone reductase {Sesbania
rostrata}, partial (54%)
Length = 652
Score = 84.0 bits (206), Expect(2) = 4e-32
Identities = 42/97 (43%), Positives = 60/97 (61%)
Frame = +1
Query: 105 KYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAI 164
K+ R L L Y+DL+LIHWP KQ + + + + F ++G SME+ RLG+ K+I
Sbjct: 361 KFFRNLGLGYVDLYLIHWPVRLKQDVEGHNFKGEDTIPFDIKGTWESMEDCYRLGIAKSI 540
Query: 165 GVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKL 201
GV+N IKKL L A I+P ++QVE++ W Q KL
Sbjct: 541 GVSNFGIKKLSMLLENAEIAPAVNQVEMNSSWHQGKL 651
Score = 71.6 bits (174), Expect(2) = 4e-32
Identities = 32/86 (37%), Positives = 59/86 (68%)
Frame = +2
Query: 23 MGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDG 82
+G ST+P + + T ++++A+K G++HFD A+ Y E+ +G A+++ + L+ ++D
Sbjct: 17 LGTSTSPSPPHEVLT-SILVDAIKIGYRHFDTASIYNTEEPLGQAVSKALELGLVKNRDE 193
Query: 83 LFVTSKVWVIDNHPELIVPALQKYLR 108
LFVTSK+W D H +L++PAL+ L+
Sbjct: 194 LFVTSKLWCTDAHHDLVLPALKSTLK 271
>TC78085 similar to GP|9927482|emb|CAC04887.1 unnamed protein product
{Medicago sativa}, partial (80%)
Length = 1030
Score = 128 bits (321), Expect = 3e-30
Identities = 87/284 (30%), Positives = 142/284 (49%), Gaps = 1/284 (0%)
Frame = +2
Query: 19 RILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIA 78
+I +GL T A ++ D + AV G++H D A Y E+ +G A+ + + ++
Sbjct: 110 KIPSVGLGTWLAAPGVVY--DAISTAVNVGYRHIDCAQIYGNEKEIGDALKKLFANGVV- 280
Query: 79 SKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVL 138
++ +++TSK+W D+ PE + A + LR LQL+YLDL+LIHWP + K G + P
Sbjct: 281 KREEMWITSKLWCTDHLPEDVPKAFDRTLRDLQLDYLDLYLIHWPVSIKNGHLTKP---- 448
Query: 139 EIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQ 198
+ +ME G +AIGV+N S+KKL L + P ++QVE+ QQ
Sbjct: 449 -----DIPSTWKAMEALYDSGKARAIGVSNFSVKKLQDLLDVGRVPPAVNQVELHPQLQQ 613
Query: 199 VKLRDFCKEKALYWQFGIIAFSPLRMGVSRGANLAMDNDIQKELDTHGKTIAQICL*W-I 257
L FCK K ++ + A+SPL G+ +N+ + + + GKT AQI L W +
Sbjct: 614 PNLHTFCKSKGVH----LSAYSPLGKGLE--SNILKNPVLHTTAEKLGKTPAQIALRWGL 775
Query: 258 YKHVNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQERLI 301
+ + + IF S+ E+ +E QER++
Sbjct: 776 QMGHSVLPKSTNTARIKENIDIFDWSIPEDLLANFNEFQQERVV 907
>TC82833 similar to PIR|T48188|T48188 aldose reductase-like protein -
Arabidopsis thaliana, partial (66%)
Length = 794
Score = 127 bits (319), Expect = 6e-30
Identities = 73/214 (34%), Positives = 117/214 (54%), Gaps = 1/214 (0%)
Frame = +1
Query: 83 LFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIME 142
LF+TSK+W D PE + PAL L+ LQL+YLDL+L+HWPF K GA + P + E+ E
Sbjct: 13 LFLTSKIWCTDLTPERVRPALNNTLQELQLDYLDLYLVHWPFLLKDGASR-PPKAGEVSE 189
Query: 143 FGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLR 202
F M+GV ME+ + L + IG+ N ++ KLDKL A + P + Q+E+ GW+ K+
Sbjct: 190 FDMEGVWREMEKLVKENLVRDIGICNFTLTKLDKLVNIAQVMPSVCQMEMHPGWRNDKML 369
Query: 203 DFCKEKALYWQFGIIAFSPLRMGVSRGANLAMDNDIQKELDTHGKTIAQICL*W-IYKHV 261
+ CK+ ++ + A+SPL G +L D + + K+ Q+ + W + +
Sbjct: 370 EACKKNGIH----VTAYSPLG-SQDGGRDLIHDQTVDRIAKKLNKSPGQVLVKWAMQRGT 534
Query: 262 NFCDEEL*QGEDATKLAIFGCSLTENDFEKISEI 295
+ + + +F L +NDF K+S+I
Sbjct: 535 SVIPKSTNPNRIKENVVVFNWELPDNDFNKLSKI 636
>TC83181 similar to PIR|T02543|T02543 aldehyde dehydrogenase homolog
At2g37770 - Arabidopsis thaliana, partial (74%)
Length = 655
Score = 120 bits (300), Expect = 9e-28
Identities = 70/204 (34%), Positives = 113/204 (55%)
Frame = +3
Query: 19 RILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIA 78
+I +GL T +++ V A+K G++ D A Y E+ +G+ + + ++
Sbjct: 30 KIPSVGLGTWQSDPGLVA--QAVAAAIKAGYRRIDCAQVYGNEKEIGSILKKLFAEGVVK 203
Query: 79 SKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVL 138
+D L++TSK+W D+ PE + AL + L LQL+Y+DL+LIHWP K+G+V + E
Sbjct: 204 RED-LWITSKLWNTDHAPEDVPLALDRTLTDLQLDYVDLYLIHWPAPMKKGSVGFKAE-- 374
Query: 139 EIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQ 198
+++ + +ME G +AIGV+N S KKL L A + P ++QVE W+Q
Sbjct: 375 NLVQPNLASTWKAMEALYDSGKARAIGVSNFSSKKLGDLLEVARVPPAVNQVECHPSWRQ 554
Query: 199 VKLRDFCKEKALYWQFGIIAFSPL 222
KLRDFC K ++ + +SPL
Sbjct: 555 DKLRDFCNSKGVH----LSGYSPL 614
>BG586880 weakly similar to GP|2792155|emb chalcone reductase {Sesbania
rostrata}, partial (51%)
Length = 783
Score = 117 bits (294), Expect = 4e-27
Identities = 68/199 (34%), Positives = 109/199 (54%), Gaps = 32/199 (16%)
Frame = +2
Query: 24 GLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGL 83
G S P + + T ++++A++ G++HFD A+ Y E+ +G A+++ + L+ ++D L
Sbjct: 20 GTSENPSPPHEVLT-SILVDAIEIGYRHFDTASVYNTEEPLGQAVSKALELGLVKNRDEL 196
Query: 84 FVTSKVWVIDNHPELIVPALQ--------------------------------KYLRTLQ 111
FVTSK+W D H +L++P+L+ K R L+
Sbjct: 197 FVTSKLWCTDAHHDLVLPSLKTTIKQAYFS*LLFFRFFYVCYFYYLSTFIFGLKLYRKLK 376
Query: 112 LEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSI 171
L+Y+DL+LIH+P KQ Y I+ +I+ F ++G +ME RLGL K+IGV+N I
Sbjct: 377 LDYVDLYLIHFPVRLKQDVEGYNIKSEDIIPFDIKGTWEAMEYCYRLGLAKSIGVSNFGI 556
Query: 172 KKLDKLFCFATISPVIDQV 190
KKL LF A I P ++QV
Sbjct: 557 KKLSMLFESAKIYPAVNQV 613
>TC87245 similar to GP|10334991|gb|AAG15839.2 NADPH-dependent mannose
6-phosphate reductase {Orobanche ramosa}, partial (98%)
Length = 1196
Score = 106 bits (265), Expect = 1e-23
Identities = 82/281 (29%), Positives = 137/281 (48%), Gaps = 13/281 (4%)
Frame = +3
Query: 39 DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPEL 98
D+++ ++K G++HFD AA Y E VG A+ E L+ +D LF+T+K+W D+
Sbjct: 108 DLIINSIKIGYRHFDCAADYKNEAEVGEALKEAFDTGLVKRED-LFITTKLWNSDHGH-- 278
Query: 99 IVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIE------VLEI-MEFGMQGV*SS 151
+V A + L+ LQL+YLDL+L+H+P AT+ V VL+I ++ +
Sbjct: 279 VVEACKDSLKKLQLDYLDLYLVHFPVATRHTGVGTTDSALGEDGVLDIDTTISLETTWHA 458
Query: 152 MEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALY 211
ME GL ++IG++N I ++ I P ++Q+E +Q+ L FC++ +
Sbjct: 459 MEGLVSSGLVRSIGISNYDIFLTRDCLAYSKIKPAVNQIETHPYFQRESLVKFCQKHGIC 638
Query: 212 WQFGIIAFSPLRMGVSR----GANLAMDNDIQKEL-DTHGKTIAQICL*W-IYKHVNFCD 265
+ A +PL + G +D I K L + + KT AQI L W I ++
Sbjct: 639 ----VTAHTPLGGAAANKEWFGTESCLDEQILKGLAEKYKKTAAQISLRWGIQRNTVVIP 806
Query: 266 EEL*QGEDATKLAIFGCSLTENDFEKISEIHQERLIKEPTK 306
+ +F L++ D E IS + +E +P K
Sbjct: 807 KTSKLERLKENFQVFDFELSKEDMELISSMDREYRTNQPAK 929
>TC82770 homologue to GP|2792155|emb|CAA11226.1 chalcone reductase {Sesbania
rostrata}, partial (68%)
Length = 681
Score = 103 bits (256), Expect = 1e-22
Identities = 62/176 (35%), Positives = 100/176 (56%), Gaps = 1/176 (0%)
Frame = +2
Query: 1 MATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAV 60
MA +P+V+L S ++ MG S +N + + ++A+K G++HFD+A+ Y
Sbjct: 8 MAGNKIPEVLLNSGHK-MPVIGMGTSVDNRPSNDVLA-SIFVDAIKVGYRHFDSASVYGT 181
Query: 61 EQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLI 120
E+++G A+A+ + LI S+D LF+TSK W D +LIVPAL+ L+ L EY+DL+LI
Sbjct: 182 EEAIGMAVAKALEQGLIKSRDELFITSKPWNTDADYDLIVPALKTTLKKLGTEYVDLYLI 361
Query: 121 HWPFATKQGAVKYPIEVLE-IMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLD 175
HWP + I E ++ F ++G +MEE + L+K L +K+D
Sbjct: 362 HWPVRLRHDLENPVIFTKEDLLPFDIEGTWKAMEECL*VRLSKVHWHMQLWYQKID 529
Score = 65.5 bits (158), Expect = 3e-11
Identities = 30/55 (54%), Positives = 41/55 (74%)
Frame = +3
Query: 157 RLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALY 211
+LGL K+IG+ N KKL KL ATI+P ++QVE++ WQQ KLR+FCKEK ++
Sbjct: 474 KLGLAKSIGICNYGTKKLTKLLETATITPAVNQVEMNPSWQQGKLREFCKEKGIH 638
>CA857968 similar to PIR|T48188|T48 aldose reductase-like protein -
Arabidopsis thaliana, partial (66%)
Length = 775
Score = 93.6 bits (231), Expect = 9e-20
Identities = 52/147 (35%), Positives = 83/147 (56%)
Frame = +2
Query: 110 LQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNL 169
LQL+YLDL+L+HWPF K GA + P + E+ EF M+GV ME+ + L + IG+ N
Sbjct: 8 LQLDYLDLYLVHWPFLLKDGASRPP-KAGEVSEFDMEGVWREMEKLVKENLVRDIGICNF 184
Query: 170 SIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPLRMGVSRG 229
++ KLDKL A + P + Q+E+ GW+ K+ + CK+ ++ + A+SPL G
Sbjct: 185 TLTKLDKLVNIAQVMPSVCQMEMHPGWRNDKMLEACKKNGIH----VTAYSPLG-SQDGG 349
Query: 230 ANLAMDNDIQKELDTHGKTIAQICL*W 256
+L D + + K+ Q+ + W
Sbjct: 350 RDLIHDQTVDRIAKKLNKSPGQVLVKW 430
>BF519680 weakly similar to GP|13160399|em aldose reductase {Digitalis
purpurea}, partial (43%)
Length = 569
Score = 84.3 bits (207), Expect = 5e-17
Identities = 44/116 (37%), Positives = 69/116 (58%)
Frame = +3
Query: 23 MGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDG 82
+GL T + ++ D V+ AVK G++H D A Y E+ +G A+ ++ +
Sbjct: 195 VGLGTWKASPGVVG--DAVVAAVKAGYRHIDCARVYDNEKEIGEALKTLFSAGVV-QRGE 365
Query: 83 LFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVL 138
+F+TSK+W+ D PE + AL + L LQL+Y+DL+LIHWPF TK G+ + EV+
Sbjct: 366 MFITSKLWISDCAPEDVSKALARTLEDLQLDYIDLYLIHWPFRTKSGSRGWDPEVM 533
>TC78086 similar to GP|9927482|emb|CAC04887.1 unnamed protein product
{Medicago sativa}, partial (42%)
Length = 480
Score = 71.6 bits (174), Expect = 4e-13
Identities = 35/96 (36%), Positives = 59/96 (61%)
Frame = +3
Query: 39 DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPEL 98
D + AV G++H D A Y E+ +G A+ + + ++ ++ +++TSK+W D+ PE
Sbjct: 138 DAISTAVNVGYRHIDCAQYYGNEKEIGDALKKLFANGVV-KREEMWITSKLWNTDHLPED 314
Query: 99 IVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYP 134
+ A + LR LQL+YLDL+LIH+P + K G + P
Sbjct: 315 VPKAFGRTLRDLQLDYLDLYLIHFPVSMKNGQLTKP 422
>BQ751060 similar to GP|14330324|emb aldo-keto reductase {Gallus gallus},
partial (20%)
Length = 827
Score = 61.2 bits (147), Expect = 5e-10
Identities = 44/145 (30%), Positives = 72/145 (49%), Gaps = 7/145 (4%)
Frame = +1
Query: 48 GFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVI-DNHPELIVPALQKY 106
G+K D A YA E VG + E + ++ +FV +K+W E + L+K
Sbjct: 259 GYKAIDGAYCYANEDEVGQGLKEAFASGV--KREDIFVVTKLWATYTTSDERVKEGLEKS 432
Query: 107 LRTLQLEYLDLFLIHWPFA--TKQGAVKYPIEVLE----IMEFGMQGV*SSMEESQRLGL 160
L++L L+Y+DL+L+HWP A + ++P + + +F S+E+ G
Sbjct: 433 LKSLGLDYVDLYLVHWPVAMNPEGNHDRFPTKPDGSRDILRDFDHVETWKSVEKLVETGK 612
Query: 161 TKAIGVNNLSIKKLDKLFCFATISP 185
++IGV N S K LD+L + I P
Sbjct: 613 VRSIGVCNYSKKYLDQLLPRSKIVP 687
Score = 27.3 bits (59), Expect = 7.9
Identities = 21/73 (28%), Positives = 34/73 (45%)
Frame = +2
Query: 150 SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKA 209
SS ++ + L A N S + A SP ++Q+E Q ++ D C+EK
Sbjct: 593 SSSRRARSVPLVSATTPRNTSTSSSPE----ARSSPPVNQIENHPSLPQQEIVDLCREKG 760
Query: 210 LYWQFGIIAFSPL 222
+ I+A+SPL
Sbjct: 761 NH----IMAYSPL 787
>BF520883 weakly similar to PIR|S78113|S78 aldehyde reductase (NADPH) (EC
1.1.1.-) - fungus (Sporidiobolus salmonicolor), partial
(30%)
Length = 449
Score = 60.5 bits (145), Expect = 8e-10
Identities = 32/86 (37%), Positives = 51/86 (59%), Gaps = 1/86 (1%)
Frame = +2
Query: 41 VLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASK-DGLFVTSKVWVIDNHPELI 99
VLEA+K G++H D A Y ++ + A+ + + K + +F+TSK+W + P+ +
Sbjct: 29 VLEALKAGYRHLDLAKVYGNQKEIAEALKKAFGGEVPNLKREDVFITSKLWNSQHRPKDV 208
Query: 100 VPALQKYLRTLQLEYLDLFLIHWPFA 125
AL L L LEYLDL+L+H+P A
Sbjct: 209 PGALDDCLAELGLEYLDLYLVHFPVA 286
>BI309035 weakly similar to PIR|C82294|C822 oxidoreductase Tas aldo/keto
reductase family VC0667 [imported] - Vibrio cholerae,
partial (27%)
Length = 570
Score = 39.7 bits (91), Expect = 0.002
Identities = 35/121 (28%), Positives = 54/121 (43%), Gaps = 18/121 (14%)
Frame = +1
Query: 21 LVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGA-AIAEEH-----QH 74
L +G T E + + ++ EA G FD+A Y V Q ++EE+ +H
Sbjct: 73 LCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKH 252
Query: 75 *LIASKDGLFVTSKV--------WVIDNHPEL----IVPALQKYLRTLQLEYLDLFLIHW 122
I +D L + +KV W+ L I A+ L +QL+Y+DL+ IHW
Sbjct: 253 RNIP-RDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHW 429
Query: 123 P 123
P
Sbjct: 430 P 432
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.333 0.144 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,597,449
Number of Sequences: 36976
Number of extensions: 106039
Number of successful extensions: 787
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 749
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 757
length of query: 316
length of database: 9,014,727
effective HSP length: 96
effective length of query: 220
effective length of database: 5,465,031
effective search space: 1202306820
effective search space used: 1202306820
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0215b.5


