

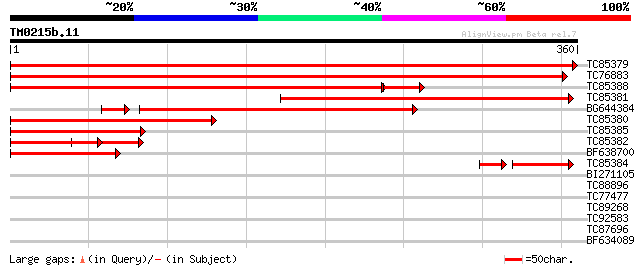
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.11
(360 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85379 similar to SP|Q43820|DCAM_PEA S-adenosylmethionine decar... 581 e-166
TC76883 homologue to SP|Q9M4D8|DCAM_VICFA S-adenosylmethionine d... 476 e-135
TC85388 similar to GP|21239731|gb|AAM44307.1 S-adenosylmethionin... 382 e-114
TC85381 similar to GP|21239731|gb|AAM44307.1 S-adenosylmethionin... 303 9e-83
BG644384 homologue to GP|807094|gb|A S-adenosylmethionine decarb... 254 3e-73
TC85380 similar to SP|Q42679|DCAM_CATRO S-adenosylmethionine dec... 223 1e-58
TC85385 weakly similar to GP|21740531|emb|CAD41510. OSJNBa0029H0... 138 3e-33
TC85382 similar to GP|21740531|emb|CAD41510. OSJNBa0029H02.26 {O... 92 3e-19
BF638700 similar to GP|21239731|gb| S-adenosylmethionine decarbo... 87 1e-17
TC85384 similar to GP|19569603|gb|AAL92115.1 hydroxyisourate hyd... 67 7e-15
BI271105 weakly similar to PIR|T46027|T46 hypothetical protein T... 30 1.4
TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown prot... 28 4.2
TC77477 homologue to GP|5305740|gb|AAD41796.1| precursor monofun... 28 4.2
TC89268 similar to PIR|T06521|T06521 pitrilysin (EC 3.4.24.55) -... 28 5.5
TC92583 similar to GP|7291175|gb|AAF46608.1| CG9817 gene product... 27 9.4
TC87696 similar to GP|9759176|dbj|BAB09791.1 contains similarity... 27 9.4
BF634089 similar to SP|Q43794|SYE_ Glutamyl-tRNA synthetase (EC ... 27 9.4
>TC85379 similar to SP|Q43820|DCAM_PEA S-adenosylmethionine decarboxylase
proenzyme (EC 4.1.1.50) (AdoMetDC) (SamDC), partial
(93%)
Length = 1774
Score = 581 bits (1498), Expect = e-166
Identities = 282/360 (78%), Positives = 319/360 (88%)
Frame = +3
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
M +T+SAIGFEGYEKRLEI+F+E G F+DP GLGLR LS+DQ+DEIL PA+CTIV SLSN
Sbjct: 501 MTMTNSAIGFEGYEKRLEITFFENGFFSDPAGLGLRALSKDQIDEILKPAECTIVDSLSN 680
Query: 61 DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF 120
D VDSYVLSESSLF+Y YK+IIKTCGTTKLLLSIP ILKLAD LNIAVKSVRYTRGSFIF
Sbjct: 681 DCVDSYVLSESSLFIYAYKLIIKTCGTTKLLLSIPAILKLADGLNIAVKSVRYTRGSFIF 860
Query: 121 PEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVY 180
P AQ FPHR+FSEEVAVL++YFG LGSG QAY+MG+ DKSQIWHIY+ASA+ + S EAVY
Sbjct: 861 PGAQSFPHRSFSEEVAVLDSYFGKLGSGSQAYMMGNADKSQIWHIYSASAKLEASPEAVY 1040
Query: 181 GLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEG 240
GLEMCMTGLDKE ASVFFK SSA LMT NSGI KILP+SDICDFEF+PCGYSMNGIEG
Sbjct: 1041GLEMCMTGLDKEKASVFFKTEGSSAGLMTKNSGIRKILPKSDICDFEFEPCGYSMNGIEG 1220
Query: 241 SAISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKL 300
SAISTIHVTPEDGFSYASFEAVGYDY + SL ELV RVLACF+PAEFS+ALH+D +GEK+
Sbjct: 1221SAISTIHVTPEDGFSYASFEAVGYDYSEKSLNELVGRVLACFYPAEFSIALHIDTNGEKI 1400
Query: 301 DKFPLDIEGYYCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCCWSEDENDEEVKGI 360
DKFPL+++GY C +R E LG GAV++ +FVR+DGC+SP+S LKCCWSEDEN+EEVK I
Sbjct: 1401DKFPLEVKGYNCGERINEVLGEDGAVVYRTFVRIDGCSSPRSTLKCCWSEDENEEEVKEI 1580
>TC76883 homologue to SP|Q9M4D8|DCAM_VICFA S-adenosylmethionine
decarboxylase proenzyme (EC 4.1.1.50) (AdoMetDC)
(SamDC), complete
Length = 1819
Score = 476 bits (1224), Expect = e-135
Identities = 231/355 (65%), Positives = 286/355 (80%), Gaps = 1/355 (0%)
Frame = +3
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
+A+ SAIGFEG+EKRLEISF + G+F+DP G GLR L++ QLDEIL PA+CTIV SL+N
Sbjct: 558 VAMAVSAIGFEGFEKRLEISFSDPGLFSDPHGRGLRSLTKSQLDEILAPAECTIVSSLAN 737
Query: 61 DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF 120
D VDSYVLSESSLFVY YK+IIKTCGTTKLLL+IP ILKLA+S+++ V+SVRYTRGSFIF
Sbjct: 738 DDVDSYVLSESSLFVYAYKIIIKTCGTTKLLLAIPPILKLAESISLKVRSVRYTRGSFIF 917
Query: 121 PEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVY 180
P AQ FPHR+FSEEVAVL+ YFG LGSG AY+MG D++Q WH+Y+ASA+ +++VY
Sbjct: 918 PGAQSFPHRHFSEEVAVLDGYFGKLGSGSTAYIMGGSDEAQNWHVYSASADSVSPNDSVY 1097
Query: 181 GLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEG 240
LEMCMTGLD+E ASVFFKD + SAA MT NSGI KILP S+ICDF+F+PCGYSMN +EG
Sbjct: 1098TLEMCMTGLDREKASVFFKDQSDSAAKMTVNSGIRKILPNSEICDFDFEPCGYSMNSVEG 1277
Query: 241 SAISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKL 300
+A+STIHVTPEDGFSYASFE GYD + ++L ELV +VLACF P EFSVA+H+D +
Sbjct: 1278AAVSTIHVTPEDGFSYASFETAGYDLKAMNLNELVVKVLACFQPNEFSVAVHVDNASKSF 1457
Query: 301 DK-FPLDIEGYYCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCCWSEDEND 354
++ LD++GY ++R E LG GG+V++ FV+ C SP+S LKC EDE +
Sbjct: 1458EQGCSLDVKGYCREERSHEGLGMGGSVVYQKFVKTADCGSPRSTLKCWKDEDEEE 1622
>TC85388 similar to GP|21239731|gb|AAM44307.1 S-adenosylmethionine
decarboxylase {x Citrofortunella mitis}, partial (69%)
Length = 1307
Score = 382 bits (981), Expect(2) = e-114
Identities = 190/239 (79%), Positives = 211/239 (87%)
Frame = +2
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
M +T+SAIGFEGYEKRLEI+F+E GVF+DP G GLR LS+DQ+DEIL PA+CTIV SLSN
Sbjct: 491 MTVTNSAIGFEGYEKRLEITFFENGVFSDPAGQGLRALSKDQIDEILKPAECTIVDSLSN 670
Query: 61 DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF 120
D VDSYVLSESSLF+Y YK+IIKTCGTTKLLLSIP ILKLAD +NIAVKSVRYTRGSFIF
Sbjct: 671 DDVDSYVLSESSLFIYAYKLIIKTCGTTKLLLSIPAILKLADCVNIAVKSVRYTRGSFIF 850
Query: 121 PEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVY 180
P AQ FPHR+FSEEVAVL++YFG LGSG QAY+MGD DKSQIWHIY+ASA+ + S EAVY
Sbjct: 851 PGAQSFPHRSFSEEVAVLDSYFGKLGSGSQAYMMGDADKSQIWHIYSASAKLEASPEAVY 1030
Query: 181 GLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIE 239
GLEMCMTGL ASVFFK + SSAA MT NSGI KILP+SDICDFEF+PCGYSMNGIE
Sbjct: 1031GLEMCMTGLR*RKASVFFKTDKSSAASMTENSGIRKILPKSDICDFEFEPCGYSMNGIE 1207
Score = 46.6 bits (109), Expect(2) = e-114
Identities = 21/26 (80%), Positives = 23/26 (87%)
Frame = +3
Query: 238 IEGSAISTIHVTPEDGFSYASFEAVG 263
++GSAISTIHVTPEDGFSYA EA G
Sbjct: 1203 LKGSAISTIHVTPEDGFSYAKLEASG 1280
>TC85381 similar to GP|21239731|gb|AAM44307.1 S-adenosylmethionine
decarboxylase {x Citrofortunella mitis}, partial (47%)
Length = 666
Score = 303 bits (775), Expect = 9e-83
Identities = 144/186 (77%), Positives = 164/186 (87%)
Frame = +3
Query: 173 KGSSEAVYGLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCG 232
+ S EAVYGLEMCMTGLDKE ASVFFK + SSA LMT NSGI KILP+SDICDFEF+PCG
Sbjct: 9 EASPEAVYGLEMCMTGLDKEKASVFFKTDASSAGLMTKNSGIRKILPKSDICDFEFEPCG 188
Query: 233 YSMNGIEGSAISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALH 292
YSMNGIEGSAISTIHVTPEDGFSYASFEAVGY++E+ SL ELV RVLACF+PAEFSVALH
Sbjct: 189 YSMNGIEGSAISTIHVTPEDGFSYASFEAVGYNFEEKSLNELVGRVLACFYPAEFSVALH 368
Query: 293 MDMHGEKLDKFPLDIEGYYCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCCWSEDE 352
+D +GEKLDKFPL+++GY C +R E LG GAV++ +FVR DGC+SP+S LKCCWSEDE
Sbjct: 369 IDTNGEKLDKFPLEVKGYNCGERINEVLGEDGAVVYRTFVRNDGCSSPRSTLKCCWSEDE 548
Query: 353 NDEEVK 358
N+EEVK
Sbjct: 549 NEEEVK 566
>BG644384 homologue to GP|807094|gb|A S-adenosylmethionine decarboxylase;
SAMDC {Solanum tuberosum}, partial (55%)
Length = 610
Score = 254 bits (648), Expect(2) = 3e-73
Identities = 121/177 (68%), Positives = 147/177 (82%)
Frame = +3
Query: 83 KTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFPEAQPFPHRNFSEEVAVLNNYF 142
K CGTTKLLL+IP IL+LA++L++ V+ VRYTRGSFIFP AQ FPHR+FSEEVAVL+ YF
Sbjct: 78 KHCGTTKLLLAIPPILRLAETLSLKVQDVRYTRGSFIFPGAQSFPHRHFSEEVAVLDGYF 257
Query: 143 GNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVYGLEMCMTGLDKESASVFFKDNT 202
G L +G +A +MG P+K+Q WH+Y+ASA P S++ VY LEMCMTGLD+E ASVF+K
Sbjct: 258 GKLAAGSKAVIMGSPNKTQKWHVYSASAGPVQSNDPVYTLEMCMTGLDREKASVFYKTEE 437
Query: 203 SSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSAISTIHVTPEDGFSYASF 259
SSAA MT SGI KILP+S+ICDFEF+PCGYSMN IE +A+STIH+TP D FSYASF
Sbjct: 438 SSAAHMTVRSGIRKILPKSEICDFEFEPCGYSMNSIERAAVSTIHITPGDRFSYASF 608
Score = 39.3 bits (90), Expect(2) = 3e-73
Identities = 18/18 (100%), Positives = 18/18 (100%)
Frame = +1
Query: 59 SNDYVDSYVLSESSLFVY 76
SNDYVDSYVLSESSLFVY
Sbjct: 4 SNDYVDSYVLSESSLFVY 57
>TC85380 similar to SP|Q42679|DCAM_CATRO S-adenosylmethionine decarboxylase
proenzyme (EC 4.1.1.50) (AdoMetDC) (SamDC), partial
(35%)
Length = 851
Score = 223 bits (567), Expect = 1e-58
Identities = 109/131 (83%), Positives = 119/131 (90%)
Frame = +1
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
M +T+SAIGFEGYEKRLEI+F+E GVF+DP GLGLR LSRDQ+DEIL PA+CTIV SLSN
Sbjct: 457 MTMTNSAIGFEGYEKRLEITFFENGVFSDPSGLGLRALSRDQIDEILKPAECTIVDSLSN 636
Query: 61 DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF 120
DYVDSYVLSESSLF+Y YK+IIKTCGTTKLLLSIP ILKLAD LNI VKSVRYTRGSFIF
Sbjct: 637 DYVDSYVLSESSLFIYAYKLIIKTCGTTKLLLSIPAILKLADGLNITVKSVRYTRGSFIF 816
Query: 121 PEAQPFPHRNF 131
P AQ FPHR+F
Sbjct: 817 PGAQSFPHRSF 849
>TC85385 weakly similar to GP|21740531|emb|CAD41510. OSJNBa0029H02.26 {Oryza
sativa}, partial (27%)
Length = 705
Score = 138 bits (348), Expect = 3e-33
Identities = 66/86 (76%), Positives = 76/86 (87%)
Frame = +1
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
MA+T+SAIGFEGYEKRLEI+F+E GVF+DP GLG R LS+DQ+DEIL PA+CTIV SLSN
Sbjct: 418 MAMTNSAIGFEGYEKRLEITFFENGVFSDPAGLGFRALSKDQIDEILKPAECTIVDSLSN 597
Query: 61 DYVDSYVLSESSLFVYPYKVIIKTCG 86
DYVDSYVLSESSLF+Y YK+ KT G
Sbjct: 598 DYVDSYVLSESSLFIYAYKLSSKTAG 675
>TC85382 similar to GP|21740531|emb|CAD41510. OSJNBa0029H02.26 {Oryza
sativa}, partial (26%)
Length = 849
Score = 92.0 bits (227), Expect = 3e-19
Identities = 43/59 (72%), Positives = 51/59 (85%)
Frame = +2
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLS 59
M +T+SAIGFEGYEKRLEI+F+E G F+DP GLGLR LS+DQ+DEIL PA+CTI SLS
Sbjct: 593 MTMTNSAIGFEGYEKRLEITFFENGFFSDPAGLGLRALSKDQIDEILKPAECTIGDSLS 769
Score = 50.4 bits (119), Expect = 1e-06
Identities = 25/46 (54%), Positives = 30/46 (64%)
Frame = +3
Query: 40 RDQLDEILNPAQCTIVGSLSNDYVDSYVLSESSLFVYPYKVIIKTC 85
+ +L N +V ND VDSYVLSESSLF+Y YK+IIKTC
Sbjct: 711 KTKLTRF*NQQNAPLVTHYLNDCVDSYVLSESSLFIYAYKLIIKTC 848
>BF638700 similar to GP|21239731|gb| S-adenosylmethionine decarboxylase {x
Citrofortunella mitis}, partial (18%)
Length = 679
Score = 86.7 bits (213), Expect = 1e-17
Identities = 44/70 (62%), Positives = 51/70 (72%)
Frame = +3
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
MAL S+IGFEG+EKRLEISF+E GVF DPEG L+ L++ Q D IL PA+C I N
Sbjct: 468 MALAVSSIGFEGFEKRLEISFFEPGVFLDPEGXDLKSLTKSQXDXILAPAECXIGFLTVN 647
Query: 61 DYVDSYVLSE 70
D DSYVLSE
Sbjct: 648 DKXDSYVLSE 677
>TC85384 similar to GP|19569603|gb|AAL92115.1 hydroxyisourate hydrolase
{Glycine max}, partial (29%)
Length = 1243
Score = 67.0 bits (162), Expect(2) = 7e-15
Identities = 28/39 (71%), Positives = 34/39 (86%)
Frame = +3
Query: 320 LGAGGAVMFHSFVRVDGCASPKSILKCCWSEDENDEEVK 358
LG GAV++ +FVR DGC+SP+S LKCCWSEDEN+EEVK
Sbjct: 66 LGEDGAVVYRTFVRNDGCSSPRSTLKCCWSEDENEEEVK 182
Score = 30.8 bits (68), Expect(2) = 7e-15
Identities = 11/17 (64%), Positives = 15/17 (87%)
Frame = +1
Query: 299 KLDKFPLDIEGYYCDKR 315
KLDKFPL+++GY C +R
Sbjct: 1 KLDKFPLEVKGYNCGER 51
>BI271105 weakly similar to PIR|T46027|T46 hypothetical protein T10K17.260 -
Arabidopsis thaliana, partial (5%)
Length = 647
Score = 30.0 bits (66), Expect = 1.4
Identities = 32/140 (22%), Positives = 57/140 (39%), Gaps = 5/140 (3%)
Frame = +2
Query: 209 TTNSGINKILPQSDICDFEFDPCGYSMNGIEGSAISTIHVTPEDGFSYASFEAVGYDYED 268
T S +N + C F D C ++ + + S+ H E + S E V E
Sbjct: 125 TGGSALNNSSDPTQSCPFSSDNCSSCLSEGDNNTTSSNHENQESSTTSDS-EDVCQQSEV 301
Query: 269 LSLTELVERVLACFHPAEFSVALHMDMHGEKLDKFPLDIEGYYCDKRGTEELGAGGAVMF 328
+ VE+VL+ H E ++ + + +GE L + + G D GT +G V
Sbjct: 302 RDNSACVEKVLSDCH--EVAMENNQNANGESLSRSSSSLTGASFD--GTRSDASGNFVEI 469
Query: 329 -----HSFVRVDGCASPKSI 343
+ F + C+ P+++
Sbjct: 470 GHSFGNGFSTTNVCSQPQNL 529
>TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown protein
{Arabidopsis thaliana}, partial (27%)
Length = 1097
Score = 28.5 bits (62), Expect = 4.2
Identities = 20/88 (22%), Positives = 40/88 (44%)
Frame = +2
Query: 165 IYAASAEPKGSSEAVYGLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDIC 224
+ AA E S+ + LE + + KE A + S++++ T NK++ + +IC
Sbjct: 386 LLAAKEENIEMSKKIESLESEIETVSKEKAQALNNEKLSASSVQTLLEEKNKLINELEIC 565
Query: 225 DFEFDPCGYSMNGIEGSAISTIHVTPED 252
E + +M+ + SA+ + D
Sbjct: 566 RDEEEKTKLAMDSL-ASALHEVSAEARD 646
>TC77477 homologue to GP|5305740|gb|AAD41796.1| precursor monofunctional
aspartokinase {Glycine max}, partial (84%)
Length = 2339
Score = 28.5 bits (62), Expect = 4.2
Identities = 19/68 (27%), Positives = 35/68 (50%), Gaps = 3/68 (4%)
Frame = +3
Query: 29 DPEGLGLRELSRDQLDEILNPAQ-CTIVGSLSNDYVDSYV--LSESSLFVYPYKVIIKTC 85
DP L REL + +LD ++ + +V L N + S + + +SSL + +++T
Sbjct: 1491 DPSKLWSRELIQQELDHVVEELEKIAVVNLLQNRSIISLIGNVQQSSLILEKAFRVLRTL 1670
Query: 86 GTTKLLLS 93
G T ++S
Sbjct: 1671 GVTVQMIS 1694
>TC89268 similar to PIR|T06521|T06521 pitrilysin (EC 3.4.24.55) - garden
pea, partial (29%)
Length = 1127
Score = 28.1 bits (61), Expect = 5.5
Identities = 27/96 (28%), Positives = 39/96 (40%)
Frame = +2
Query: 123 AQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVYGL 182
AQP P RNF E + F + SG Q + D + Y A P V G
Sbjct: 695 AQPRPLRNFKSEQEFIPPSFRSSSSGLQFQEVFLNDTDERACAYIAGPAPNRWGFTVDGK 874
Query: 183 EMCMTGLDKESASVFFKDNTSSAALMTTNSGINKIL 218
++ T +D S+ DN + + + T G+ K L
Sbjct: 875 DLLET-IDNASS---VNDNGTKSDAVPTEGGLQKSL 970
>TC92583 similar to GP|7291175|gb|AAF46608.1| CG9817 gene product
{Drosophila melanogaster}, partial (1%)
Length = 1015
Score = 27.3 bits (59), Expect = 9.4
Identities = 18/38 (47%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Frame = -2
Query: 321 GAGGAVMFHSFVRVDGCASPK-SILKCCWSEDENDEEV 357
G GG V+ +VRV G ASP S CC S+ EEV
Sbjct: 354 GWGGGVISGFWVRVAGSASPDVSAHGCCMSQVVWQEEV 241
>TC87696 similar to GP|9759176|dbj|BAB09791.1 contains similarity to
pathogenesis-related genes transcriptional
activator~gene_id:K19E1.9, partial (24%)
Length = 1725
Score = 27.3 bits (59), Expect = 9.4
Identities = 34/110 (30%), Positives = 46/110 (40%), Gaps = 13/110 (11%)
Frame = +3
Query: 210 TNSGINKILPQSDI-CDFEFDP---CGYSMNGIEGSAIST--------IHVTPEDGFSYA 257
TN I ++ +++I DFE P G S G A+S+ TP DGF Y
Sbjct: 1044 TNYPITQVEVKTEIEKDFEVTPPVSSGNSDRGGYSDALSSPTSVLPYDCDSTPFDGFRYV 1223
Query: 258 SFEAVGYDYE-DLSLTELVERVLACFHPAEFSVALHMDMHGEKLDKFPLD 306
+A G+ + LSL E V L C E EK ++F LD
Sbjct: 1224 DVDAFGFHIDAPLSLPE-VNVPLTCHQKLE---------KEEKFEEFDLD 1343
>BF634089 similar to SP|Q43794|SYE_ Glutamyl-tRNA synthetase (EC 6.1.1.17)
(Glutamate--tRNA ligase) (GluRS). [Common tobacco],
partial (22%)
Length = 427
Score = 27.3 bits (59), Expect = 9.4
Identities = 10/29 (34%), Positives = 17/29 (58%)
Frame = -2
Query: 274 LVERVLACFHPAEFSVALHMDMHGEKLDK 302
++E + HP+ FS+ H D +KLD+
Sbjct: 231 ILEAIFLV*HPSSFSLVPHQDRSSDKLDR 145
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,250,250
Number of Sequences: 36976
Number of extensions: 154620
Number of successful extensions: 707
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 681
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 706
length of query: 360
length of database: 9,014,727
effective HSP length: 97
effective length of query: 263
effective length of database: 5,428,055
effective search space: 1427578465
effective search space used: 1427578465
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0215b.11


