

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0211.14
(280 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
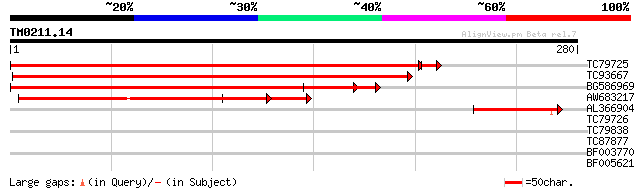
Score E
Sequences producing significant alignments: (bits) Value
TC79725 similar to GP|6729027|gb|AAF27023.1| unknown protein {Ar... 390 e-109
TC93667 similar to GP|6729027|gb|AAF27023.1| unknown protein {Ar... 328 1e-90
BG586969 similar to GP|6729027|gb| unknown protein {Arabidopsis ... 271 2e-73
AW683217 similar to GP|6069661|dbj Similar to Sequence of BAC F7... 135 2e-32
AL366904 weakly similar to GP|6729027|gb|A unknown protein {Arab... 84 8e-17
TC79726 similar to GP|17064808|gb|AAL32558.1 Unknown protein {Ar... 35 0.042
TC79838 similar to GP|8164030|gb|AAF73960.1| SGS3 {Arabidopsis t... 31 0.46
TC87877 similar to GP|21618275|gb|AAM67325.1 unknown {Arabidopsi... 31 0.60
BF003770 similar to PIR|F96809|F96 protein F28K19.26 [imported] ... 30 1.0
BF005621 27 6.7
>TC79725 similar to GP|6729027|gb|AAF27023.1| unknown protein {Arabidopsis
thaliana}, partial (63%)
Length = 760
Score = 390 bits (1001), Expect(2) = e-109
Identities = 184/204 (90%), Positives = 196/204 (95%)
Frame = +3
Query: 1 MGVLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCS 60
MGVLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTP GI+QAR AG L R+++ GCS
Sbjct: 114 MGVLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPSGIAQARHAGANLHRLVSGQGCS 293
Query: 61 PDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQT 120
PDWR+ FYVSPYARTRSTLREVGRSFSKKRVIGVREESR+REQDFGNFQV+ERMKI+K+T
Sbjct: 294 PDWRLYFYVSPYARTRSTLREVGRSFSKKRVIGVREESRVREQDFGNFQVQERMKIVKET 473
Query: 121 RERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSR 180
RERFGRFFYRFPEGESAADV+DRISGFFE+LWRDIDLNRLHHDP+NDLNLVIVSHGLTSR
Sbjct: 474 RERFGRFFYRFPEGESAADVFDRISGFFESLWRDIDLNRLHHDPTNDLNLVIVSHGLTSR 653
Query: 181 IFLMKWFKWTVEQFEHLNNFGNCE 204
IFLMKWFKWTVEQFEHLNNFGNCE
Sbjct: 654 IFLMKWFKWTVEQFEHLNNFGNCE 725
Score = 22.7 bits (47), Expect(2) = e-109
Identities = 9/10 (90%), Positives = 10/10 (100%)
Frame = +1
Query: 204 EFRVMEQGSG 213
+FRVMEQGSG
Sbjct: 724 KFRVMEQGSG 753
>TC93667 similar to GP|6729027|gb|AAF27023.1| unknown protein {Arabidopsis
thaliana}, partial (58%)
Length = 662
Score = 328 bits (842), Expect = 1e-90
Identities = 153/198 (77%), Positives = 176/198 (88%)
Frame = +2
Query: 2 GVLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSP 61
GVLPKRII++RHGESQGNLD AYT TPDH I LTPQGISQA G+R+R V+++ SP
Sbjct: 68 GVLPKRIIVVRHGESQGNLDPGAYTVTPDHKIPLTPQGISQALSTGSRIRHVISSSSSSP 247
Query: 62 DWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTR 121
DWRV FYVSPY RTRSTLRE+ +SFSKKRVIGVREE RIREQDFGNFQV+ERM IK+TR
Sbjct: 248 DWRVYFYVSPYTRTRSTLRELAKSFSKKRVIGVREECRIREQDFGNFQVQERMDAIKETR 427
Query: 122 ERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRI 181
+RFGRFFYRFPEGESAADV+DR+S F E++WRDID+NRL+H+PSNDLNL+IVSHGL SR+
Sbjct: 428 QRFGRFFYRFPEGESAADVFDRVSSFLESMWRDIDMNRLNHNPSNDLNLIIVSHGLASRV 607
Query: 182 FLMKWFKWTVEQFEHLNN 199
FLM+WF+WTVEQFE LNN
Sbjct: 608 FLMRWFRWTVEQFELLNN 661
>BG586969 similar to GP|6729027|gb| unknown protein {Arabidopsis thaliana},
partial (56%)
Length = 788
Score = 271 bits (693), Expect = 2e-73
Identities = 135/172 (78%), Positives = 151/172 (87%)
Frame = +1
Query: 1 MGVLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCS 60
MGVLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTP GI+QAR AG L R+++ GCS
Sbjct: 154 MGVLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPSGIAQARHAGANLHRLVSGQGCS 333
Query: 61 PDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQT 120
PDWR+ FYVSPYARTRSTLREVGRSFSKKRVIGVREESR+REQDFGNFQV+ERMKI+K+T
Sbjct: 334 PDWRLYFYVSPYARTRSTLREVGRSFSKKRVIGVREESRVREQDFGNFQVQERMKIVKET 513
Query: 121 RERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVI 172
RERFGRFFYRFPEGESAADV+DRISG L+ + + ++HH N LNL +
Sbjct: 514 RERFGRFFYRFPEGESAADVFDRISGNC*LLFPSL*IIQIHHIDIN-LNLFL 666
Score = 80.5 bits (197), Expect = 7e-16
Identities = 36/38 (94%), Positives = 38/38 (99%)
Frame = +3
Query: 146 GFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIFL 183
GFFE+LWRDIDLNRLHHDP+NDLNLVIVSHGLTSRIFL
Sbjct: 675 GFFESLWRDIDLNRLHHDPTNDLNLVIVSHGLTSRIFL 788
>AW683217 similar to GP|6069661|dbj Similar to Sequence of BAC F7G19 from
Arabidopsis thaliana chromosome 1; similar to
Saccharomyces, partial (29%)
Length = 664
Score = 135 bits (340), Expect = 2e-32
Identities = 66/125 (52%), Positives = 84/125 (66%)
Frame = +2
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+RIIL+RHGES+GN+D S YT PD I LT +G QA G R++ ++ D +W+
Sbjct: 230 PRRIILVRHGESEGNVDESVYTRVPDPKIGLTNRGRVQAEECGQRIKNMIEKDS-DENWQ 406
Query: 65 VNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERF 124
+ FYVSPY RT TL+ + R F + R+ G REE RIREQDFGNFQ E MK+ K R +
Sbjct: 407 LYFYVSPYRRTLETLQSLARPFERSRIAGFREEPRIREQDFGNFQNRELMKVEKAQRHLY 586
Query: 125 GRFFY 129
GRFFY
Sbjct: 587 GRFFY 601
Score = 42.0 bits (97), Expect = 3e-04
Identities = 23/44 (52%), Positives = 28/44 (63%)
Frame = +3
Query: 106 GNFQVEERMKIIKQTRERFGRFFYRFPEGESAADVYDRISGFFE 149
G F++E K+ K+ F RFP GESAADVYDRI+GF E
Sbjct: 531 GIFKIES**KLKKRNAIFMVVSFTRFPNGESAADVYDRITGFRE 662
>AL366904 weakly similar to GP|6729027|gb|A unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 407
Score = 83.6 bits (205), Expect = 8e-17
Identities = 35/45 (77%), Positives = 39/45 (85%), Gaps = 1/45 (2%)
Frame = +3
Query: 230 WGLSPEMVADQKWRAGASRGAWNDQCSWYLDGFFDRL-AEESDDE 273
WG+SP+M+ADQKWRA A RGAWNDQC WYLDGFFDR A ESDD+
Sbjct: 3 WGMSPDMIADQKWRADAPRGAWNDQCPWYLDGFFDRFDASESDDD 137
>TC79726 similar to GP|17064808|gb|AAL32558.1 Unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 777
Score = 34.7 bits (78), Expect = 0.042
Identities = 33/127 (25%), Positives = 60/127 (46%), Gaps = 1/127 (0%)
Frame = +3
Query: 6 KRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWRV 65
KR++L+RHG+S N + + D S+ LT +G SQA + R+++ D
Sbjct: 255 KRVVLVRHGQSTWNAE-GRIQGSSDFSV-LTKKGESQAETS----RQMLLEDNFDA---- 404
Query: 66 NFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQT-RERF 124
+ SP AR++ T + + G R++ I E D + ++K+ + RF
Sbjct: 405 -CFASPLARSKKT---------AEIIWGSRQQQIIPEYDLREIDLYSFQGLLKEEGKARF 554
Query: 125 GRFFYRF 131
G F+++
Sbjct: 555 GPAFHQW 575
>TC79838 similar to GP|8164030|gb|AAF73960.1| SGS3 {Arabidopsis thaliana},
partial (28%)
Length = 1287
Score = 31.2 bits (69), Expect = 0.46
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = +1
Query: 86 FSKKRVIGVREESRIREQDFGNFQVEERMKIIKQT 120
F K+R+ + ++ +E+DF Q EER K+ K T
Sbjct: 655 FFKERISSIHDKRNAKEEDFERMQQEEREKVKKTT 759
>TC87877 similar to GP|21618275|gb|AAM67325.1 unknown {Arabidopsis
thaliana}, partial (79%)
Length = 1387
Score = 30.8 bits (68), Expect = 0.60
Identities = 45/212 (21%), Positives = 86/212 (40%), Gaps = 26/212 (12%)
Frame = +2
Query: 8 IILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRV-------------- 53
+IL+RHGES N + + +T D + L+ +GI +A AG R+ +
Sbjct: 569 LILIRHGESLWN-EKNLFTGCVD--VPLSKKGIDEAIEAGKRISSIPVDLIFTSALIRAQ 739
Query: 54 ---MAADGCSPDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQV 110
M A +V + + +V +KK+ I V ++ E+ +G Q
Sbjct: 740 MTAMLAMTQHRRRKVPIVMHNESEQAKAWSQVFSEDTKKQSIPVIAAWQLNERMYGELQG 919
Query: 111 EERMKIIKQTRERFGR---------FFYRFPEGESAADVYDRISGFFEALWRDIDLNRLH 161
+ ++T +R+G+ + P GES +R +F +++
Sbjct: 920 LNK----QETADRYGKEQVHEWRRSYDIPPPNGESLEMCAERAVAYFR--------DQIE 1063
Query: 162 HDPSNDLNLVIVSHGLTSRIFLMKWFKWTVEQ 193
+ N++I +HG + R +M K T ++
Sbjct: 1064PQLLSGKNVMIAAHGNSLRSIIMYLDKLTSQE 1159
>BF003770 similar to PIR|F96809|F96 protein F28K19.26 [imported] -
Arabidopsis thaliana, partial (15%)
Length = 570
Score = 30.0 bits (66), Expect = 1.0
Identities = 17/43 (39%), Positives = 27/43 (62%)
Frame = +3
Query: 8 IILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRL 50
+IL+RHGES N + + +T D + LT +G+ +A AG R+
Sbjct: 246 LILIRHGESMWN-EKNLFTGCCD--VPLTRKGVEEAIEAGKRI 365
>BF005621
Length = 464
Score = 27.3 bits (59), Expect = 6.7
Identities = 20/76 (26%), Positives = 40/76 (52%), Gaps = 3/76 (3%)
Frame = +1
Query: 88 KKRVIGVREESRIREQDFGNFQVEERMKIIKQTR-ERFGRFFYR--FPEGESAADVYDRI 144
+ R + +REE++ +Q F ER+K+ ++ ER R + F + E + ++
Sbjct: 202 ENRYMRLREENQKLQQSFEM----ERLKLREEFHLERLSRVYAESEFKKREEICEKGKKV 369
Query: 145 SGFFEALWRDIDLNRL 160
+EAL +++ +NRL
Sbjct: 370 KESYEALLKEVKVNRL 417
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,953,829
Number of Sequences: 36976
Number of extensions: 121303
Number of successful extensions: 589
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 585
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 587
length of query: 280
length of database: 9,014,727
effective HSP length: 95
effective length of query: 185
effective length of database: 5,502,007
effective search space: 1017871295
effective search space used: 1017871295
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0211.14


