

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
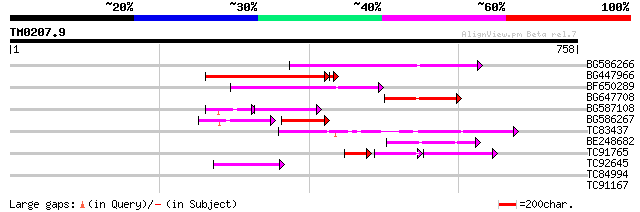
Query= TM0207.9
(758 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 198 7e-51
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 153 4e-37
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 132 6e-31
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 109 4e-24
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 71 2e-18
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 61 4e-18
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 79 6e-15
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 63 4e-10
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 57 3e-08
TC92645 49 5e-06
TC84994 homologue to SP|P28553|CRTI_SOYBN Phytoene dehydrogenase... 31 2.0
TC91167 weakly similar to GP|20147231|gb|AAM10330.1 At1g68570/F2... 29 7.7
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 198 bits (503), Expect = 7e-51
Identities = 102/258 (39%), Positives = 156/258 (59%)
Frame = -3
Query: 375 THFRPISLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMK 434
+ +R I+ CN +KII L+ +++ +L IIS QSAFVPGR ++DN L+ ++ H+++
Sbjct: 775 SEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYLR 596
Query: 435 KKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNG 494
+ + MA+K DM+KAYDR+ W FLR L ++GF W+S+IM+ V+TV + ++NG
Sbjct: 595 QSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLING 416
Query: 495 NPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHL 554
PQ P RGLRQGDPLSPYLF++C EV S L Q+ + L G+K+ARN P I+HL
Sbjct: 415 GPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARN--CPPINHL 242
Query: 555 LFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLL 614
LFADD + F ++ + + I+ +Y + IN KS +++S + +K L
Sbjct: 241 LFADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGEL 62
Query: 615 GVKAMDNYDRYLGLPTII 632
+ +YLG I+
Sbjct: 61 KIAKEGGTGKYLGYRNIL 8
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 153 bits (387), Expect(2) = 4e-37
Identities = 80/166 (48%), Positives = 104/166 (62%)
Frame = +2
Query: 262 LMDIFSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGL 321
L+ F+ LFT+SNP IE +V +L+ H+ F+ E+V E + QMHP KA G
Sbjct: 140 LITYFNNLFTSSNPTAIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGP 319
Query: 322 DGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPIS 381
DGL LF+QK+WHIVG +V L VL + + T +VLIPK K +RPIS
Sbjct: 320 DGLPALFFQKYWHIVGKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPIS 499
Query: 382 LCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAY 427
LCNV KIIT +AN++K LPD+I QSAFV GRL+TDNAL+A+
Sbjct: 500 LCNVVMKIITKVIANRVKQTLPDVIDVEQSAFVQGRLITDNALIAW 637
Score = 20.4 bits (41), Expect(2) = 4e-37
Identities = 7/12 (58%), Positives = 9/12 (74%)
Frame = +1
Query: 428 ECFHFMKKKIYG 439
ECFH++K K G
Sbjct: 637 ECFHWLKXKKKG 672
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 132 bits (331), Expect = 6e-31
Identities = 69/204 (33%), Positives = 118/204 (57%)
Frame = +3
Query: 296 ILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPG 355
+L + F+ +V+ LF M +KA G+DG + F++ W+I+GD V L + P
Sbjct: 9 LLCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPK 188
Query: 356 TIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVP 415
I T + L+PK +FRPI+ C+V +KII+ L ++++ VL ++S QSAFV
Sbjct: 189 IINCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVK 368
Query: 416 GRLVTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNW 475
GR++ DN ++++E +K G + +K+D+ KAYD EWPF++ + ++GFP +
Sbjct: 369 GRVIFDNIILSHELVKSYSRK--GISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKF 542
Query: 476 VSFIMDYVTTVLFQVMLNGNPQVP 499
V+++M +TT + NG+ P
Sbjct: 543 VNWVMAXLTTASYTFNXNGDLTXP 614
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 109 bits (272), Expect = 4e-24
Identities = 53/103 (51%), Positives = 75/103 (72%)
Frame = +1
Query: 502 PGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNV 561
P +GLRQGDPLSPYLF++C V S L++R+ +L GI++AR S P I+HLLFADD++
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVAR--SDPKITHLLFADDSL 177
Query: 562 IFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPS 604
+FARA EA+ + +L Y Q++NF+KS +SYS+NVP+
Sbjct: 178 LFARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPN 306
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 71.2 bits (173), Expect(2) = 2e-18
Identities = 39/89 (43%), Positives = 50/89 (55%)
Frame = +3
Query: 328 FYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNVPF 387
F+Q WHI+ D+ S L + T + LIPK K+ T RPISLCNV +
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 388 KIIT*TLANKLKIVLPDIISGPQSAFVPG 416
KII+ L +LK+ LP +IS QSAFV G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
Score = 40.0 bits (92), Expect(2) = 2e-18
Identities = 25/71 (35%), Positives = 37/71 (51%), Gaps = 4/71 (5%)
Frame = +2
Query: 263 MDIFSTLFTTSNPMG----IEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKA 318
+D F LF + P G ++ TS + ++N+ L + E+VR LF MHP KA
Sbjct: 215 VDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLATE----EEVRLALFIMHPEKA 382
Query: 319 SGLDGLHTLFY 329
G DG+ TL +
Sbjct: 383 PGPDGMTTLLF 415
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 60.8 bits (146), Expect(2) = 4e-18
Identities = 30/64 (46%), Positives = 45/64 (69%)
Frame = +1
Query: 364 LIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNA 423
L+PK ++ FRPISLCNV +KI++ L+ +LK VLP II+ Q+AF +L++DN
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 424 LVAY 427
L+A+
Sbjct: 781 LIAH 792
Score = 49.3 bits (116), Expect(2) = 4e-18
Identities = 33/109 (30%), Positives = 52/109 (47%), Gaps = 6/109 (5%)
Frame = +3
Query: 253 WRMRTYPGFLMDIFSTLFTTSNPMGI-----EAATSLVANRLNKGHLAILNTPFSGEKVR 307
W + G L D L +S +GI + ++V N A L S E+VR
Sbjct: 264 WFVEEDLGRLADSHFKLLYSSEDVGITLEDWNSIPAIVTEEQN----AQLMAQISREEVR 431
Query: 308 EPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSVLQ-GRVSPG 355
E +F ++P K G DG++ F+Q+FW +GDD+ L+ G++ G
Sbjct: 432 EAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEG 578
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 79.0 bits (193), Expect = 6e-15
Identities = 82/326 (25%), Positives = 138/326 (42%), Gaps = 5/326 (1%)
Frame = +2
Query: 360 TLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLV 419
T + LIPK+ FRPISL +KI+ LAN+L++V+ +IS QSAFV R +
Sbjct: 59 TFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKNRQI 238
Query: 420 TDNALVAYECFHFMK----KKIYGC-NGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVN 474
+ + + +M+ +KI+ C I+ + +S + W M F V
Sbjct: 239 LEMVFL-*QMRLWMRLRN*RKIFCCLRWILKRLITLSIG---LIWILF*VG---MSFLVL 397
Query: 475 WVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGA 534
W +I + V+T V++NG+P + +L+Q ++
Sbjct: 398 WRKWIKECVSTATTSVLVNGSP-----------------------TNVLMKSLVQTQLFT 508
Query: 535 SHLSGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKS 594
+ G+ V+ V+SHL FA+D ++ ++ L + + +NF KS
Sbjct: 509 RYSFGV-----VNPVVVSHLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVNFHKS 673
Query: 595 MLSYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGR 654
L PS + +E +L K YLG+P + + + R +L G
Sbjct: 674 GLVCVNIAPS-WLSEAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPIVNRIKARLTGW 850
Query: 655 KEPSLSRAGREVLIKAVAQEIPAYIM 680
LS GR VL+K+V + Y +
Sbjct: 851 NSRFLSFGGRLVLLKSVLTSLSVYAL 928
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 63.2 bits (152), Expect = 4e-10
Identities = 40/126 (31%), Positives = 61/126 (47%)
Frame = +3
Query: 504 RGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIF 563
RGL+QGDPL+P+LFL+ E S L++ + + G + R + +SHL +ADD +
Sbjct: 24 RGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTR--VSHLQYADDTLCI 197
Query: 564 ARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYD 623
T + +K +L ++ +NF KS L NVP + L +
Sbjct: 198 GMPTVDNLWTLKALLQGFEMASGLKVNFHKSSL-IGINVPRDFMEAACRFLNCREESIPF 374
Query: 624 RYLGLP 629
YLGLP
Sbjct: 375 IYLGLP 392
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 57.0 bits (136), Expect = 3e-08
Identities = 34/99 (34%), Positives = 52/99 (52%)
Frame = +3
Query: 554 LLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKML 613
LLF + R A +K IL Y++ + I+ KS + SRNVP + + +
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 614 LGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLK 652
LGV+ M +YLGLP++IG+ +T F +K W+K K
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTFSSIKGGVWQKNK 575
Score = 46.2 bits (108), Expect(2) = 6e-08
Identities = 27/65 (41%), Positives = 35/65 (53%)
Frame = +2
Query: 488 FQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVS 547
+ V++N + P P RGL+QGD LSPY+F+IC E S LI G I R
Sbjct: 122 YYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTSI*R--G 295
Query: 548 APVIS 552
AP +S
Sbjct: 296 APPVS 310
Score = 29.3 bits (64), Expect(2) = 6e-08
Identities = 15/36 (41%), Positives = 23/36 (63%)
Frame = +1
Query: 448 LDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYV 483
L +SK Y+RV+ +L+ + KMGF W+ + M YV
Sbjct: 1 LYISKVYNRVD*DYLKEIMIKMGFNNRWI-YWMFYV 105
>TC92645
Length = 561
Score = 49.3 bits (116), Expect = 5e-06
Identities = 31/96 (32%), Positives = 49/96 (50%), Gaps = 1/96 (1%)
Frame = +3
Query: 273 SNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFYQKF 332
+N ++V ++ + +L PFS +++ +FQMH KA GLDGL+ Y +F
Sbjct: 234 ANSSNFSIVLNVVCRKIGDQNNCMLTAPFSVDEIWTAIFQMHHDKAFGLDGLNLAVYYRF 413
Query: 333 WHIVGDDVAHFCLSVLQG-RVSPGTIIPTLLVLIPK 367
++VG D + L L G R T +VL+PK
Sbjct: 414 *NLVGGDTNYVGLYSLVG*R*VA*LYWGTNIVLVPK 521
>TC84994 homologue to SP|P28553|CRTI_SOYBN Phytoene dehydrogenase
chloroplast precursor (EC 1.14.99.-) (Phytoene
desaturase). [Soybean], partial (25%)
Length = 757
Score = 30.8 bits (68), Expect = 2.0
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 4/38 (10%)
Frame = -2
Query: 112 ICFGLRRFGSKLVRNVQKLWQ----RHGTQEVLILTQK 145
+CFGL++F V VQ LW RH + +L L QK
Sbjct: 180 LCFGLQKFHQGRVWQVQTLWHR*YLRHYVKSILRLVQK 67
>TC91167 weakly similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7
{Arabidopsis thaliana}, partial (5%)
Length = 728
Score = 28.9 bits (63), Expect = 7.7
Identities = 13/40 (32%), Positives = 22/40 (54%)
Frame = +2
Query: 461 FLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPF 500
F+ SAL+ MGF N VS ++ ++ + F + + N F
Sbjct: 332 FVLSALDNMGFVTNMVSLVLYFIGVMHFDLSSSANTLTNF 451
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.334 0.146 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,847,952
Number of Sequences: 36976
Number of extensions: 338157
Number of successful extensions: 2639
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1465
Number of HSP's successfully gapped in prelim test: 143
Number of HSP's that attempted gapping in prelim test: 1141
Number of HSP's gapped (non-prelim): 1663
length of query: 758
length of database: 9,014,727
effective HSP length: 103
effective length of query: 655
effective length of database: 5,206,199
effective search space: 3410060345
effective search space used: 3410060345
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0207.9


