

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
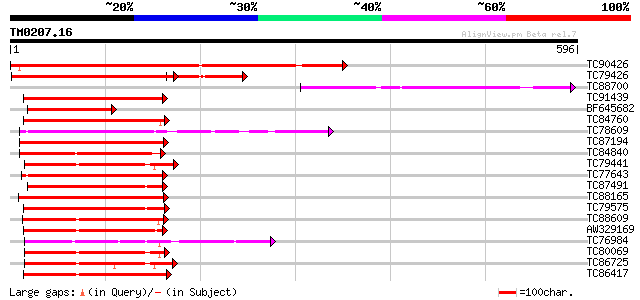
Query= TM0207.16
(596 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90426 similar to GP|10177859|dbj|BAB11211. NAC2-like protein {... 469 e-132
TC79426 similar to GP|7270509|emb|CAB80274.1 NAM / CUC2-like pro... 328 3e-90
TC88700 240 1e-63
TC91439 similar to PIR|D96683|D96683 hypothetical protein F12P19... 197 7e-51
BF645682 similar to GP|7270509|emb| NAM / CUC2-like protein {Ara... 182 3e-46
TC84760 similar to GP|8567777|gb|AAF76349.1| unknown protein {Ar... 176 2e-44
TC78609 similar to GP|21105746|gb|AAM34772.1 nam-like protein 9 ... 176 2e-44
TC87194 NAC1 175 5e-44
TC84840 similar to PIR|C84671|C84671 NAM (no apical meristem)-li... 174 1e-43
TC79441 similar to GP|9757865|dbj|BAB08499.1 NAM (no apical meri... 172 3e-43
TC77643 similar to GP|21105742|gb|AAM34770.1 nam-like protein 7 ... 172 4e-43
TC87491 similar to PIR|T48437|T48437 hypothetical protein T32M21... 170 1e-42
TC88165 similar to GP|7716952|gb|AAF68626.1| NAC1 {Medicago trun... 167 1e-41
TC79575 similar to GP|7715611|gb|AAF68129.1| F20B17.1 {Arabidops... 161 8e-40
TC88609 similar to GP|15148914|gb|AAK84884.1 NAC domain protein ... 160 1e-39
AW329169 similar to PIR|E84636|E84 NAM (no apical meristem)-like... 160 2e-39
TC76984 similar to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like... 159 4e-39
TC80069 similar to GP|15148914|gb|AAK84884.1 NAC domain protein ... 158 6e-39
TC86725 similar to GP|9758909|dbj|BAB09485.1 NAM (no apical meri... 157 1e-38
TC86417 similar to GP|16226943|gb|AAL16305.1 AT4g27410/F27G19_10... 157 1e-38
>TC90426 similar to GP|10177859|dbj|BAB11211. NAC2-like protein {Arabidopsis
thaliana}, partial (36%)
Length = 1234
Score = 469 bits (1207), Expect = e-132
Identities = 237/368 (64%), Positives = 288/368 (77%), Gaps = 13/368 (3%)
Frame = +3
Query: 1 MGAVETAVP---------VLSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREI 51
MGAV +P VLSLN LP GFRFRP+DEE++DFYLR KINGNGDEVWVIRE+
Sbjct: 141 MGAVVECLPSPQQAEELAVLSLNKLPLGFRFRPTDEELVDFYLRMKINGNGDEVWVIREV 320
Query: 52 DVCKWEPWDLPDLSVIRNRDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSG 111
DVCK+EPWDLPDLS++RN+DPEWFFFCP DRKYPNG+RLNRAT GYWKATGKDRKIKSG
Sbjct: 321 DVCKFEPWDLPDLSIVRNKDPEWFFFCPMDRKYPNGSRLNRATTNGYWKATGKDRKIKSG 500
Query: 112 SSLIGMKKTLVFYHGRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESI 171
++LIGMKKTLVFY GRAPKG+RT+W+MHEYRPTL+ELDGTNPGQNPYV+CRLFKKQDES+
Sbjct: 501 ATLIGMKKTLVFYAGRAPKGQRTHWIMHEYRPTLKELDGTNPGQNPYVLCRLFKKQDESL 680
Query: 172 EVSISGEAEQTASTPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTS 231
S GEAEQT STP A YSPEE+QSD L LV +SSSL TED++H A IPE SEE S
Sbjct: 681 ASSNCGEAEQTTSTPTTANYSPEEIQSD--LNLVPLSSSLATEDERHLAAIPENSEEAMS 854
Query: 232 NIITPVDRNSDRYDAQNAQPQNVKL-AAEENQPLNLDMYNDLKNDI--FDDKLFSPALVH 288
++IT D SD +A +AQ Q++++ AAEE+Q LNLD+++ + ++ +DDKLFSPA H
Sbjct: 855 HVITTADCYSDACNASDAQHQSLEVPAAEEDQLLNLDIFDSPRFELEPWDDKLFSPAHAH 1034
Query: 289 LPPVFDYQASNEPDDRSGLQYGTNETAISDLFDS-LTWDQLSYEESGSQPMNFPLFNVKD 347
PP F +QA+NE +QYGTN T +SD FDS + WD+ E S S ++ +F + +
Sbjct: 1035FPPEFGHQANNEL-----IQYGTNGTDVSDFFDSCVNWDEFFSEASISLELSPNIFKIPE 1199
Query: 348 SGSGSDSD 355
+GS S+SD
Sbjct: 1200NGSCSNSD 1223
>TC79426 similar to GP|7270509|emb|CAB80274.1 NAM / CUC2-like protein
{Arabidopsis thaliana}, partial (28%)
Length = 1011
Score = 328 bits (842), Expect = 3e-90
Identities = 149/175 (85%), Positives = 163/175 (93%)
Frame = +1
Query: 3 AVETAVPVLSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLP 62
+V+ + VLSLNSLP GFRFRP+DEE+ID+YLR KINGNGDEVWVIREIDVCKWEPWD+P
Sbjct: 268 SVDDGMAVLSLNSLPLGFRFRPTDEELIDYYLRSKINGNGDEVWVIREIDVCKWEPWDMP 447
Query: 63 DLSVIRNRDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLV 122
DLSV+RN+DPEWFFFCPQDRKYPNG+RLNRAT GYWKATGKDRKIKSG+ LIGMKKTLV
Sbjct: 448 DLSVVRNKDPEWFFFCPQDRKYPNGHRLNRATNHGYWKATGKDRKIKSGTILIGMKKTLV 627
Query: 123 FYHGRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISG 177
FY GRAPKGKRTNWVMHEYRPTL+ELDGTNPGQNPYV+CRLFKK DES+EVS G
Sbjct: 628 FYSGRAPKGKRTNWVMHEYRPTLKELDGTNPGQNPYVLCRLFKKNDESVEVSNYG 792
Score = 89.4 bits (220), Expect = 4e-18
Identities = 46/86 (53%), Positives = 61/86 (70%)
Frame = +2
Query: 165 KKQDESIEVSISGEAEQTASTPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPE 224
+K +++ GE EQT S P+AA YSPEE+QSD P P + VS+S +TE+DK A+IP+
Sbjct: 755 RKTMRALKFQTMGEVEQTTSAPLAANYSPEEIQSD-PAP-ITVSTSQVTEEDKQLAVIPD 928
Query: 225 TSEETTSNIITPVDRNSDRYDAQNAQ 250
SEET SN+IT VD +SD YDA + Q
Sbjct: 929 ISEETISNVITSVDYHSDGYDAHDVQ 1006
>TC88700
Length = 1080
Score = 240 bits (613), Expect = 1e-63
Identities = 147/293 (50%), Positives = 174/293 (59%), Gaps = 4/293 (1%)
Frame = +3
Query: 306 GLQYGTNETAISDLFD-SLTWDQLSYEESGSQPMNFPLFNVKDSGSGSDSDFEITNMPSM 364
GLQYGTNE ISD + L WDQ+ E S S NFP+ NVKDSGSGS+SD E+ NM M
Sbjct: 3 GLQYGTNEINISDFLNYDLNWDQIPCEVSVSHQPNFPMLNVKDSGSGSNSDVEVANMTGM 182
Query: 365 QIPYPEEAIDMKIPLGTAPEFFSPVGVPFDHSGDEQKSNVGLFQNQNAAQMTFSSDV-SM 423
Q YP EAID +IP T P F FD+SGD+QKSNV L QN Q F SDV +
Sbjct: 183 QAAYPHEAIDRRIPFATTPSF----SHTFDYSGDDQKSNVVLLQNN--FQTAFPSDVNNT 344
Query: 424 GQVYNVVNDYEQPRNWNVATGGDTGIIIRARQAQNELVNTNINQGSANRRIRLLKSAHGS 483
G+VYN VN YEQPR N ++GI+ RAR ++E TN QG+A RRIRL K H S
Sbjct: 345 GEVYNAVNGYEQPRTCNTIMSSESGIVRRARPTRDEQFKTNPMQGTAQRRIRLSKIEHVS 524
Query: 484 SRVVKGGRRA--QEDRNSKPITAVEKKASESLAADEDDTLTNHVKAMPKTPKSTNNRKIY 541
+VK + QE NSKPI AVEK A + DE TNH K PKS N
Sbjct: 525 CGMVKVKDESCTQEQHNSKPIIAVEKAAENHASDDESAICTNHACKQRKVPKSIAN---- 692
Query: 542 QRGSNAGSSTKVLKDLLLLRRVPCISKASSNRAMWSSVFVVSAFVMVSVMVFS 594
++K L++ LLL R P SK SSN A+WSSVFVVSA V+VS+ F+
Sbjct: 693 --------ASKGLRNFLLLGRAPFRSKTSSNCALWSSVFVVSASVLVSLAAFT 827
>TC91439 similar to PIR|D96683|D96683 hypothetical protein F12P19.8
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1154
Score = 197 bits (502), Expect = 7e-51
Identities = 89/152 (58%), Positives = 114/152 (74%)
Frame = +3
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SLP GFRF P+DEE++ +YL +KING+ E+ +I E+D+ K EPWDLP S++ D EW
Sbjct: 48 SLPPGFRFHPTDEELVAYYLNRKINGHKIELEIIAEVDLYKCEPWDLPGKSLLPGNDMEW 227
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF P+DRKYPNG+R NRAT GYWKATGKDRK+ S + +G+KKTLV+Y GRAP G RT
Sbjct: 228 YFFSPRDRKYPNGSRTNRATKSGYWKATGKDRKVNSQARAVGVKKTLVYYRGRAPHGSRT 407
Query: 135 NWVMHEYRPTLQELDGTNPGQNPYVICRLFKK 166
NWVMHEYR +E + T Q+ Y +CR+ KK
Sbjct: 408 NWVMHEYRLDERECETTPSLQDAYALCRVIKK 503
>BF645682 similar to GP|7270509|emb| NAM / CUC2-like protein {Arabidopsis
thaliana}, partial (17%)
Length = 668
Score = 182 bits (462), Expect = 3e-46
Identities = 80/94 (85%), Positives = 88/94 (93%)
Frame = +3
Query: 19 GFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWFFFC 78
GFRFRP+DEE+ID+YLR KINGNGDEV VIREIDVCKWEPWD+PDLSVI++ DPEWFFFC
Sbjct: 387 GFRFRPTDEELIDYYLRLKINGNGDEVQVIREIDVCKWEPWDMPDLSVIKDNDPEWFFFC 566
Query: 79 PQDRKYPNGNRLNRATILGYWKATGKDRKIKSGS 112
PQDR+YPNGNRLNR T GYWKATGKDRKIKSG+
Sbjct: 567 PQDRQYPNGNRLNRETNHGYWKATGKDRKIKSGT 668
>TC84760 similar to GP|8567777|gb|AAF76349.1| unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 781
Score = 176 bits (446), Expect = 2e-44
Identities = 82/160 (51%), Positives = 108/160 (67%), Gaps = 6/160 (3%)
Frame = +2
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SL GFRF P+DEE++ +YL++K++G I E+D+ K EPWDL D S ++ RD EW
Sbjct: 215 SLAPGFRFHPTDEELVIYYLKRKVSGKPFRFDAIAEVDIYKSEPWDLFDKSRLKTRDQEW 394
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF D+KY NG R+NRAT+ GYWKATG DR +K +G+KKTLVF+ GRAP GKRT
Sbjct: 395 YFFSALDKKYGNGGRMNRATLQGYWKATGNDRPVKHDLRTVGLKKTLVFHSGRAPDGKRT 574
Query: 135 NWVMHEYRPTLQELDGTNPGQNP------YVICRLFKKQD 168
NWVMHEYR +EL+ G P +V+C +F K +
Sbjct: 575 NWVMHEYRLVEEELERERAGAGPSQPQEAFVLCXVFHKNN 694
>TC78609 similar to GP|21105746|gb|AAM34772.1 nam-like protein 9 {Petunia x
hybrida}, partial (39%)
Length = 1931
Score = 176 bits (446), Expect = 2e-44
Identities = 123/336 (36%), Positives = 175/336 (51%), Gaps = 6/336 (1%)
Frame = +1
Query: 11 LSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWV-IREIDVCKWEPWDLPDLSVIRN 69
+SL P GFRF P+DEEI+ FYL++K+ G + + ID+ K+EPWDLP++S ++
Sbjct: 103 MSLYQAP-GFRFHPTDEEIVCFYLKRKLTGKLPPCFDHLAFIDIYKFEPWDLPNMSKLKT 279
Query: 70 RDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAP 129
+D EW+FF D+KY G R NRAT GYWK TGKDR IK+G +GMKKTLV++ GRAP
Sbjct: 280 KDLEWYFFSALDKKYGTGCRTNRATDRGYWKTTGKDRVIKNGEETVGMKKTLVYHLGRAP 459
Query: 130 KGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAA 189
G R+NWVMHEYR +EL T GQ+ YV+CR+F+K SG + A
Sbjct: 460 HGNRSNWVMHEYRMIDEELAKTG-GQDAYVLCRIFEK---------SGSGPKNGEKYGAP 609
Query: 190 KYSPEEVQSDTPL-PLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNSDRYDAQN 248
E D P+ PL V + LL ++ A P+ +E S + D +
Sbjct: 610 FVEAEWQNVDEPVNPLPPVDNQLL---EQVAAPAPDYAEFLQSLVAFDNDLLN------- 759
Query: 249 AQPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHLPPVFDY-QASNEPD-DRSG 306
++V A ++N LD+ +L D + LPP F Y + S+ P +S
Sbjct: 760 ---ESVPTATDDNYLEILDLDQEL------DTSVTVGNADLPPNFYYGECSSFPQHSQSE 912
Query: 307 LQYGTNETAISDLF--DSLTWDQLSYEESGSQPMNF 340
+G + + L+ DS+ P+NF
Sbjct: 913 AAFGIYDHQVGGLYGVDSVQDGYNGETNHNENPLNF 1020
>TC87194 NAC1
Length = 2067
Score = 175 bits (443), Expect = 5e-44
Identities = 77/157 (49%), Positives = 115/157 (73%)
Frame = +2
Query: 11 LSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNR 70
+++ L GFRF P+ E+I ++L++K+ G VI E+D+ K+ PWDLPDLS ++N
Sbjct: 443 MAIPKLVPGFRFSPTGVELIKYFLKRKVMGKKFLNDVIAELDIYKYAPWDLPDLSYLQNG 622
Query: 71 DPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPK 130
D EW+FFCP ++KY NG+R+NRAT +G+WKATGKDR ++ + +GM KTL+F+ G+AP+
Sbjct: 623 DLEWYFFCPIEKKYGNGSRMNRATEIGFWKATGKDRAVQQNNQTVGMIKTLIFHTGKAPR 802
Query: 131 GKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQ 167
G RT+WVMHEYR ++L Q+ YVIC++F+K+
Sbjct: 803 GDRTDWVMHEYRLEDKDLADKGIVQDSYVICKVFQKE 913
>TC84840 similar to PIR|C84671|C84671 NAM (no apical meristem)-like protein
[imported] - Arabidopsis thaliana, partial (47%)
Length = 683
Score = 174 bits (440), Expect = 1e-43
Identities = 82/153 (53%), Positives = 114/153 (73%)
Frame = +1
Query: 11 LSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNR 70
++ +S+ GFRF P+DEE+I +YLR+K++G+ + V VI E+++CK+EPWDLP S I++
Sbjct: 217 IAASSMFPGFRFCPTDEELISYYLRKKLDGHEESVQVISEVELCKYEPWDLPAKSFIQS- 393
Query: 71 DPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPK 130
D EWFFF P+ RKYPNG++ RAT GYWKATGK+R +KSGS++IG K+TLVF+ RAPK
Sbjct: 394 DNEWFFFSPRGRKYPNGSQSKRATDCGYWKATGKERNVKSGSNVIGTKRTLVFHLXRAPK 573
Query: 131 GKRTNWVMHEYRPTLQELDGTNPGQNPYVICRL 163
G+RT W+MHEY + + D V+CRL
Sbjct: 574 GERTEWIMHEYCVSDRSPDS-------LVVCRL 651
>TC79441 similar to GP|9757865|dbj|BAB08499.1 NAM (no apical meristem)-like
protein {Arabidopsis thaliana}, partial (61%)
Length = 1247
Score = 172 bits (437), Expect = 3e-43
Identities = 87/169 (51%), Positives = 112/169 (65%), Gaps = 7/169 (4%)
Frame = +3
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE+I YL +K+ + I ++D+ K EPWDLP + + + EW+
Sbjct: 189 LPPGFRFHPTDEELISHYLYKKVIDSNFSARAIGDVDLNKSEPWDLPFKAKMGEK--EWY 362
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FFC +DRKYP G R NRAT GYWKATGKD++I G SL+GMKKTLVFY GRAPKG+++N
Sbjct: 363 FFCVRDRKYPTGLRTNRATEAGYWKATGKDKEIFKGKSLVGMKKTLVFYKGRAPKGEKSN 542
Query: 136 WVMHEYRPTLQELDG-------TNPGQNPYVICRLFKKQDESIEVSISG 177
WVMHEYR L+G +N +VICR+F+K + ISG
Sbjct: 543 WVMHEYR-----LEGKFSVHNLPKAAKNEWVICRVFQKSSAGKKTHISG 674
>TC77643 similar to GP|21105742|gb|AAM34770.1 nam-like protein 7 {Petunia x
hybrida}, partial (27%)
Length = 1611
Score = 172 bits (435), Expect = 4e-43
Identities = 82/154 (53%), Positives = 110/154 (71%)
Frame = +2
Query: 13 LNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDP 72
++S+P GFRF P+DEE++ +YL++KI G + VI+E DV KW+P DLP+ S ++ D
Sbjct: 617 MSSMP-GFRFHPTDEELVMYYLKRKICGKKLKFNVIKETDVYKWDPEDLPEQSFLKTGDR 793
Query: 73 EWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGK 132
+WFFFC +DRKYPNG R +RAT GYWKATGKDR + S +G+KKTLVFY GRAP G+
Sbjct: 794 QWFFFCHRDRKYPNGGRSSRATRHGYWKATGKDRNVIYNSRSVGVKKTLVFYLGRAPSGE 973
Query: 133 RTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKK 166
RT+WVMHEY EL ++ Y + ++FKK
Sbjct: 974 RTDWVMHEYTMDEDELRTCRDVKDYYALYKVFKK 1075
>TC87491 similar to PIR|T48437|T48437 hypothetical protein T32M21.10 -
Arabidopsis thaliana, partial (29%)
Length = 857
Score = 170 bits (431), Expect = 1e-42
Identities = 81/149 (54%), Positives = 105/149 (70%), Gaps = 1/149 (0%)
Frame = +1
Query: 19 GFRFRPSDEEIIDFYLRQKINGNGDEVW-VIREIDVCKWEPWDLPDLSVIRNRDPEWFFF 77
GFRF P+DEEI+ +YL++K+ GN + + ID+ K+EPWDLP LS ++ RD EW+FF
Sbjct: 136 GFRFHPTDEEIVRYYLKKKLTGNLPSCFDYLAVIDIYKFEPWDLPKLSKLKTRDLEWYFF 315
Query: 78 CPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTNWV 137
D+KY N R NRAT GYWK TGKDR IK+G +GMKKTLV++ GRAP G R+NWV
Sbjct: 316 TALDKKYGNSIRTNRATDRGYWKTTGKDRAIKNGEETVGMKKTLVYHSGRAPHGDRSNWV 495
Query: 138 MHEYRPTLQELDGTNPGQNPYVICRLFKK 166
MHEYR + E Q+ YV+CR+F+K
Sbjct: 496 MHEYR-MIDETLAKAGVQDAYVLCRIFEK 579
>TC88165 similar to GP|7716952|gb|AAF68626.1| NAC1 {Medicago truncatula},
partial (61%)
Length = 1231
Score = 167 bits (423), Expect = 1e-41
Identities = 72/158 (45%), Positives = 114/158 (71%)
Frame = +1
Query: 10 VLSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRN 69
++++ L GFRF P+ E+I ++L++K+ G VI E+D+ K+ PWDLP+LS ++N
Sbjct: 460 LMAIPKLVPGFRFSPTGVELIKYFLKRKVLGKKFHNDVIAELDIYKYAPWDLPELSYLQN 639
Query: 70 RDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAP 129
D EW+FFCP ++KY +G R+ RAT +G+WKATG+DR ++ + +GM KTL+F+ G++P
Sbjct: 640 GDLEWYFFCPIEKKYRSGPRMKRATEIGFWKATGQDRAVQQNNQTVGMIKTLIFHTGKSP 819
Query: 130 KGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQ 167
+G RT+WVMHEYR ++L Q+ YVIC++F+K+
Sbjct: 820 RGDRTDWVMHEYRLEDKDLADKGTVQDSYVICKVFQKE 933
>TC79575 similar to GP|7715611|gb|AAF68129.1| F20B17.1 {Arabidopsis
thaliana}, partial (58%)
Length = 1324
Score = 161 bits (407), Expect = 8e-40
Identities = 77/156 (49%), Positives = 110/156 (70%), Gaps = 2/156 (1%)
Frame = +2
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVI-RNRDPE 73
++P GFRF P+DEE++ +YLR+K++ ++ VIRE+D+ + EPWDL D I E
Sbjct: 125 TVPPGFRFHPTDEELLYYYLRKKVSYEAIDLDVIREVDLNRLEPWDLKDKCRIGSGPQNE 304
Query: 74 WFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKI-KSGSSLIGMKKTLVFYHGRAPKGK 132
W+FF +D+KYP G R NRAT G+WKATG+D+ I + S IGM+KTLVFY GRAP G
Sbjct: 305 WYFFSHKDKKYPTGTRTNRATTAGFWKATGRDKAIYHTNSKRIGMRKTLVFYTGRAPHGH 484
Query: 133 RTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQD 168
+T+W+MHEYR L + D ++ +V+CR+FKK++
Sbjct: 485 KTDWIMHEYR--LDDDDNAEVQEDGWVVCRVFKKKN 586
>TC88609 similar to GP|15148914|gb|AAK84884.1 NAC domain protein NAC2
{Phaseolus vulgaris}, partial (71%)
Length = 1076
Score = 160 bits (406), Expect = 1e-39
Identities = 79/158 (50%), Positives = 106/158 (67%), Gaps = 4/158 (2%)
Frame = +1
Query: 14 NSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPE 73
+ LP GFRF P+DEE+I YL + +I E+D+ K +PW+LPD S E
Sbjct: 25 SELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKCDPWELPDKSEFEEN--E 198
Query: 74 WFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKR 133
W+FF P++RKYPNG R NRAT+ GYWKATG D+ IKSGS IG+KK+LVFY GR PKG +
Sbjct: 199 WYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVK 378
Query: 134 TNWVMHEYRPT-LQELDGTNPGQ---NPYVICRLFKKQ 167
T+W+MHEYR Q+ + G + +V+CR++KK+
Sbjct: 379 TDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKK 492
>AW329169 similar to PIR|E84636|E84 NAM (no apical meristem)-like protein
[imported] - Arabidopsis thaliana, partial (52%)
Length = 631
Score = 160 bits (404), Expect = 2e-39
Identities = 83/154 (53%), Positives = 106/154 (67%), Gaps = 2/154 (1%)
Frame = +3
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
+LP GFRF P+DEE+I YL KI+ + I +ID+ K EPW+LP + + + EW
Sbjct: 30 TLPPGFRFHPTDEELITCYLINKISDSSFTGKAITDIDLNKSEPWELPGKAKMGEK--EW 203
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSG--SSLIGMKKTLVFYHGRAPKGK 132
+FF +DRKYP G R NRAT GYWK TGKDR+I S L+GMKKTLVFY GRAP+G+
Sbjct: 204 YFFNMRDRKYPTGVRTNRATNTGYWKTTGKDREIFDSVTSELVGMKKTLVFYKGRAPRGE 383
Query: 133 RTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKK 166
++NWVMHEYR + TN Q+ +VICR+FKK
Sbjct: 384 KSNWVMHEYRIHSKSTFRTNK-QDEWVICRIFKK 482
>TC76984 similar to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like protein
{Arabidopsis thaliana}, partial (71%)
Length = 1450
Score = 159 bits (401), Expect = 4e-39
Identities = 97/273 (35%), Positives = 151/273 (54%), Gaps = 9/273 (3%)
Frame = +2
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE+++ YL +K G V V++E+D+ K++PW LP++ + + EW+
Sbjct: 155 LPPGFRFHPTDEELVNHYLCRKCAGQSISVPVVKEVDLYKFDPWQLPEMGY--HSEKEWY 328
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FF P+DRKYPNG+R NRA GYWKATG D+ I + G+KK LVFY G+APKG +TN
Sbjct: 329 FFSPRDRKYPNGSRPNRAAGSGYWKATGADKPIGKLKPM-GIKKALVFYAGKAPKGVKTN 505
Query: 136 WVMHEYRPTLQELDGTNPGQN-----PYVICRLFKKQDESIEVSISGEAEQTASTPMAAK 190
W+MHEYR L +D + +N +V+CR++ K+ + E T++TP
Sbjct: 506 WIMHEYR--LANVDRSAGKKNNLRLDDWVLCRIYNKKGKI-------EKFNTSNTPKVQN 658
Query: 191 -YSPEEVQSDTPLPLVAVSSSLLTEDDKHQAI--IPETSEETTSNIITPVDRNSDRYDAQ 247
Y EEV+ P + + D ++ + S + ++++P D DR
Sbjct: 659 YYEHEEVEEHEMKPEIQKLGNYQLYMDTSDSVPKLHTDSSSSGGHVVSP-DVTCDREVQS 835
Query: 248 NAQPQNVKLAAEENQPLNLD-MYNDLKNDIFDD 279
+ ++ + ++ L+ + N L DI DD
Sbjct: 836 EPKWNDLGIQLDDAFDFQLNYLENALSFDIDDD 934
>TC80069 similar to GP|15148914|gb|AAK84884.1 NAC domain protein NAC2
{Phaseolus vulgaris}, partial (63%)
Length = 1058
Score = 158 bits (399), Expect = 6e-39
Identities = 79/162 (48%), Positives = 105/162 (64%), Gaps = 9/162 (5%)
Frame = +2
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE+I +YL + +I E+D+ K++PW+LPD S EW+
Sbjct: 137 LPPGFRFHPTDEELIVYYLCNQATSQPCPASIIPEVDIYKFDPWELPDKSGFGEN--EWY 310
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FF P++RKYPNG R NRAT+ GYWKATG D+ I SGS IG+KK LVFY GR PKG +T+
Sbjct: 311 FFTPRERKYPNGVRPNRATVSGYWKATGTDKSIFSGSKHIGVKKALVFYKGRPPKGIKTD 490
Query: 136 WVMHEYRPTLQELDGTNPGQN---------PYVICRLFKKQD 168
W+MHEYR L G+ N +V+CR++KK++
Sbjct: 491 WIMHEYR-----LIGSRRQANRQIGSMRLDDWVLCRIYKKKN 601
>TC86725 similar to GP|9758909|dbj|BAB09485.1 NAM (no apical meristem)-like
protein {Arabidopsis thaliana}, partial (50%)
Length = 1645
Score = 157 bits (396), Expect = 1e-38
Identities = 84/172 (48%), Positives = 109/172 (62%), Gaps = 11/172 (6%)
Frame = +3
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+D EII YL +K+ + I E D+ K EPWDLP + + + EW+
Sbjct: 177 LPPGFRFHPTDAEIIVCYLTEKVKNSKFSATAIGEADLNKCEPWDLPKKAKMGEK--EWY 350
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKI----KSGSSLIGMKKTLVFYHGRAPKG 131
FFC +DRKYP G R NRAT GYWKATGKD++I K +L+GMKKTLVFY GRAPKG
Sbjct: 351 FFCQKDRKYPTGMRTNRATESGYWKATGKDKEIYHKGKGIQNLVGMKKTLVFYKGRAPKG 530
Query: 132 KRTNWVMHEYRPTLQELDG-------TNPGQNPYVICRLFKKQDESIEVSIS 176
++TNWVMHE+R L+G N ++ +V+ R+F K + + IS
Sbjct: 531 EKTNWVMHEFR-----LEGKFATHNLPNKEKDEWVVSRVFHKNTDVKKPQIS 671
>TC86417 similar to GP|16226943|gb|AAL16305.1 AT4g27410/F27G19_10
{Arabidopsis thaliana}, partial (67%)
Length = 1628
Score = 157 bits (396), Expect = 1e-38
Identities = 73/156 (46%), Positives = 103/156 (65%)
Frame = +2
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SLP GFRF P+DEE++ YL +K+ G+ + +I EID+ K++PW LP ++ + EW
Sbjct: 173 SLPPGFRFYPTDEELLVQYLCRKVAGHHFSLQIIAEIDLYKFDPWILPSKAIFGEK--EW 346
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF P+DRKYPNG R NR GYWKATG D+ I + +G+KK LVFY G+APKG +T
Sbjct: 347 YFFSPRDRKYPNGTRPNRVAGSGYWKATGTDKIITNEGRKVGIKKALVFYVGKAPKGTKT 526
Query: 135 NWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDES 170
NW+MHEYR + + +V+CR++KK +
Sbjct: 527 NWIMHEYRLLDSSRNNGGTKLDDWVLCRIYKKNSSA 634
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.131 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,571,580
Number of Sequences: 36976
Number of extensions: 221850
Number of successful extensions: 943
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 860
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 862
length of query: 596
length of database: 9,014,727
effective HSP length: 102
effective length of query: 494
effective length of database: 5,243,175
effective search space: 2590128450
effective search space used: 2590128450
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0207.16


