

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
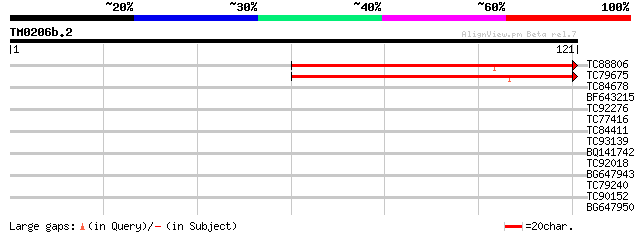
Query= TM0206b.2
(121 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88806 similar to PIR|T52589|T52589 ribose-phosphate pyrophosph... 86 4e-18
TC79675 similar to GP|16604386|gb|AAL24199.1 At1g10700/F20B24.13... 75 6e-15
TC84678 similar to GP|15529228|gb|AAK97708.1 At1g03950/F21M11_12... 30 0.22
BF643215 similar to GP|15529228|gb At1g03950/F21M11_12 {Arabidop... 30 0.22
TC92276 similar to GP|15529228|gb|AAK97708.1 At1g03950/F21M11_12... 27 1.9
TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl g... 26 3.2
TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersico... 26 3.2
TC93139 weakly similar to GP|12056928|gb|AAG48132.1 putative res... 26 4.2
BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvo... 26 4.2
TC92018 similar to PIR|D84651|D84651 hypothetical protein At2g25... 26 4.2
BG647943 similar to GP|12081917|dbj cytosolic cysteine synthase ... 25 7.1
TC79240 similar to GP|8777426|dbj|BAA97016.1 contains similarity... 25 7.1
TC90152 homologue to GP|13278055|gb|AAH03883.1 Similar to RIKEN ... 25 9.3
BG647950 similar to GP|21928129|gb| AT4g17080/dl4570w {Arabidops... 25 9.3
>TC88806 similar to PIR|T52589|T52589 ribose-phosphate pyrophosphokinase (EC
2.7.6.1) [imported] - Arabidopsis thaliana, partial
(85%)
Length = 1226
Score = 85.5 bits (210), Expect = 4e-18
Identities = 47/62 (75%), Positives = 49/62 (78%), Gaps = 1/62 (1%)
Frame = +2
Query: 61 FLAQLSSPTQGFEQFTAIYALHCLFVASFALVPPFFPTGSFE-*KEEGDVTTAFTLARML 119
FLA SSP Q FEQ + IYAL LFVASF LV PFFPTGSFE +EEGDV TAFTLARML
Sbjct: 212 FLASFSSPAQVFEQLSVIYALPRLFVASFTLVLPFFPTGSFERMEEEGDVATAFTLARML 391
Query: 120 SN 121
SN
Sbjct: 392 SN 397
>TC79675 similar to GP|16604386|gb|AAL24199.1 At1g10700/F20B24.13
{Arabidopsis thaliana}, partial (45%)
Length = 907
Score = 75.1 bits (183), Expect = 6e-15
Identities = 42/62 (67%), Positives = 45/62 (71%), Gaps = 1/62 (1%)
Frame = +3
Query: 61 FLAQLSSPTQGFEQFTAIYALHCLFVASFALVPPFFPTGSFE*KE-EGDVTTAFTLARML 119
FLA SSP FEQ IYAL LFVASF LV PFFPTG+ E E EGD+ TAFTLAR+L
Sbjct: 522 FLASFSSPAVIFEQIPVIYALPKLFVASFTLVLPFFPTGTSERMEDEGDIATAFTLARLL 701
Query: 120 SN 121
SN
Sbjct: 702 SN 707
>TC84678 similar to GP|15529228|gb|AAK97708.1 At1g03950/F21M11_12
{Arabidopsis thaliana}, partial (94%)
Length = 607
Score = 30.0 bits (66), Expect = 0.22
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = -1
Query: 83 CLFVASFALVPPFFPTGSFE 102
CLF+A+ ALV PF PT + E
Sbjct: 349 CLFIAAIALVAPFIPTATEE 290
>BF643215 similar to GP|15529228|gb At1g03950/F21M11_12 {Arabidopsis
thaliana}, partial (58%)
Length = 616
Score = 30.0 bits (66), Expect = 0.22
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = -3
Query: 83 CLFVASFALVPPFFPTGSFE 102
CLF+A+ ALV PF PT + E
Sbjct: 317 CLFIAAIALVAPFIPTATEE 258
>TC92276 similar to GP|15529228|gb|AAK97708.1 At1g03950/F21M11_12
{Arabidopsis thaliana}, partial (91%)
Length = 880
Score = 26.9 bits (58), Expect = 1.9
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = -2
Query: 83 CLFVASFALVPPFFP 97
CLF+A+ ALV PF P
Sbjct: 387 CLFIATIALVAPFIP 343
>TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl glucosamine
transferase {Arabidopsis thaliana}, partial (94%)
Length = 3465
Score = 26.2 bits (56), Expect = 3.2
Identities = 12/29 (41%), Positives = 15/29 (51%)
Frame = +2
Query: 79 YALHCLFVASFALVPPFFPTGSFE*KEEG 107
YA HC +AS +PPF K+EG
Sbjct: 1907 YAAHCSVIASRFSLPPFSHPAPIPIKQEG 1993
>TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersicon
esculentum}, partial (38%)
Length = 804
Score = 26.2 bits (56), Expect = 3.2
Identities = 10/27 (37%), Positives = 15/27 (55%)
Frame = +2
Query: 62 LAQLSSPTQGFEQFTAIYALHCLFVAS 88
L LSS T+ + +Y +HC F+ S
Sbjct: 455 LGSLSSSTKNHQMLCGLYVVHCRFLIS 535
>TC93139 weakly similar to GP|12056928|gb|AAG48132.1 putative resistance
protein {Glycine max}, partial (3%)
Length = 611
Score = 25.8 bits (55), Expect = 4.2
Identities = 15/50 (30%), Positives = 28/50 (56%)
Frame = -1
Query: 43 VSKSMYE*CRRVVW*TCCFLAQLSSPTQGFEQFTAIYALHCLFVASFALV 92
+S S+ * ++ W TCC LA+ ++ Q ++FT I H + + + L+
Sbjct: 182 ISNSLSG*INKIFWRTCC-LAECTA-IQKIKRFTFIQLFHSINMVNSMLL 39
>BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvox carteri,
partial (9%)
Length = 1337
Score = 25.8 bits (55), Expect = 4.2
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Frame = +3
Query: 65 LSSPTQGFEQFTAIYALHCLFVASFAL-VPPFFPTGSF 101
LSSP F +A C F++SF + VPPFFP F
Sbjct: 297 LSSPLLFFISPSA-----CFFLSSFPMRVPPFFPLPFF 395
>TC92018 similar to PIR|D84651|D84651 hypothetical protein At2g25680
[imported] - Arabidopsis thaliana, partial (38%)
Length = 946
Score = 25.8 bits (55), Expect = 4.2
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = -1
Query: 7 KGGFW*SIWN 16
KGGFW SIWN
Sbjct: 79 KGGFWFSIWN 50
>BG647943 similar to GP|12081917|dbj cytosolic cysteine synthase {Solanum
tuberosum}, partial (26%)
Length = 753
Score = 25.0 bits (53), Expect = 7.1
Identities = 12/33 (36%), Positives = 17/33 (51%)
Frame = +2
Query: 58 TCCFLAQLSSPTQGFEQFTAIYALHCLFVASFA 90
+C F LSS + F ++ CL VA+FA
Sbjct: 143 SCAFFLMLSSKSNASF*FIKTFSSDCLHVATFA 241
>TC79240 similar to GP|8777426|dbj|BAA97016.1 contains similarity to
alpha/beta hydrolase~gene_id:K9P8.9 {Arabidopsis
thaliana}, partial (35%)
Length = 1065
Score = 25.0 bits (53), Expect = 7.1
Identities = 8/20 (40%), Positives = 13/20 (65%)
Frame = -1
Query: 78 IYALHCLFVASFALVPPFFP 97
++ HCL +A ++ PPF P
Sbjct: 630 LFGSHCLLLAHCSMQPPFHP 571
>TC90152 homologue to GP|13278055|gb|AAH03883.1 Similar to RIKEN cDNA
2010107D16 gene {Mus musculus}, partial (59%)
Length = 698
Score = 24.6 bits (52), Expect = 9.3
Identities = 13/35 (37%), Positives = 18/35 (51%)
Frame = -2
Query: 85 FVASFALVPPFFPTGSFE*KEEGDVTTAFTLARML 119
FVA +PP FP SF+ E TT ++ +L
Sbjct: 694 FVALSKQLPPQFPANSFQVHEVAITTTVASILLIL 590
>BG647950 similar to GP|21928129|gb| AT4g17080/dl4570w {Arabidopsis
thaliana}, partial (11%)
Length = 353
Score = 24.6 bits (52), Expect = 9.3
Identities = 14/40 (35%), Positives = 19/40 (47%), Gaps = 1/40 (2%)
Frame = +1
Query: 59 CCFLAQLSSPT-QGFEQFTAIYALHCLFVASFALVPPFFP 97
CCFL ++ P+ + F LFV SF +P FP
Sbjct: 46 CCFLLNIALPSLRSFPLHLKFA*NSFLFVVSFTNLPLLFP 165
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.370 0.164 0.657
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,457,944
Number of Sequences: 36976
Number of extensions: 66488
Number of successful extensions: 807
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 800
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 805
length of query: 121
length of database: 9,014,727
effective HSP length: 84
effective length of query: 37
effective length of database: 5,908,743
effective search space: 218623491
effective search space used: 218623491
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0206b.2


