

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0204.4
(368 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
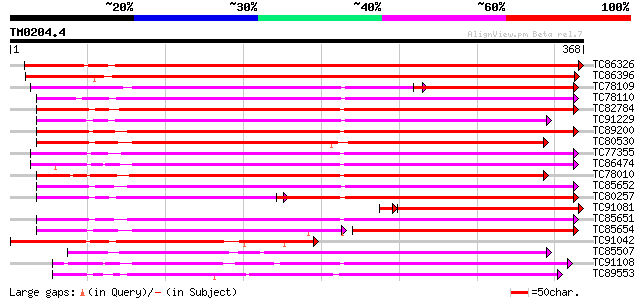
Score E
Sequences producing significant alignments: (bits) Value
TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoantho... 582 e-166
TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin ... 504 e-143
TC78109 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 179 2e-66
TC78110 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 245 2e-65
TC82784 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 241 3e-64
TC91229 similar to GP|9294689|dbj|BAB03055.1 gene_id:MHC9.10~sim... 240 6e-64
TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 239 1e-63
TC80530 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabi... 237 6e-63
TC77355 weakly similar to PIR|T05552|T05552 SRG1 protein-related... 236 1e-62
TC86474 weakly similar to GP|6016680|gb|AAF01507.1| putative leu... 234 5e-62
TC78010 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 231 3e-61
TC85652 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 229 1e-60
TC80257 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 154 1e-58
TC91081 similar to GP|5813796|gb|AAD52015.1| unknown {Pisum sati... 215 2e-58
TC85651 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 221 5e-58
TC85654 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 140 5e-58
TC91042 similar to PIR|T49209|T49209 leucoanthocyanidin dioxygen... 209 1e-54
TC85507 homologue to SP|P31239|ACCO_PEA 1-aminocyclopropane-1-ca... 209 1e-54
TC91108 similar to PIR|T05552|T05552 SRG1 protein-related protei... 207 7e-54
TC89553 weakly similar to SP|P24397|HY6H_HYONI Hyoscyamine 6-dio... 206 1e-53
>TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoanthocyanidin
dioxygenase {Arabidopsis thaliana}, partial (81%)
Length = 1686
Score = 582 bits (1499), Expect = e-166
Identities = 282/359 (78%), Positives = 312/359 (86%)
Frame = +2
Query: 10 NWPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDP 69
+WPEPI+RVQSLSE C DSIP+RYIKP TDRP +++SS D NIPIIDL GL +
Sbjct: 269 DWPEPIIRVQSLSEGCKDSIPDRYIKPPTDRPIVDTSSY--DDINIPIIDLGGL----NG 430
Query: 70 DHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSP 129
D + AS L+QIS+AC++WGFFQIVNHGVSPDLMD ARETWR+FFHLPME KQQYANSP
Sbjct: 431 DDLDVHASILKQISDACRDWGFFQIVNHGVSPDLMDKARETWRQFFHLPMEAKQQYANSP 610
Query: 130 KTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKL 189
TYEGYGSRLGVEKGAILDWSDYYFLHYLP+S+KD NKWPA P SCREVFDEYG+ELVKL
Sbjct: 611 TTYEGYGSRLGVEKGAILDWSDYYFLHYLPVSVKDCNKWPASPQSCREVFDEYGKELVKL 790
Query: 190 CGRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTL 249
GRLMK LSLNL LEE LQN FGGE+IGAC+RVNFYPKCP+PELTLGLSSHSDPGGMT+
Sbjct: 791 SGRLMKALSLNLGLEEKILQNAFGGEEIGACMRVNFYPKCPRPELTLGLSSHSDPGGMTM 970
Query: 250 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 309
LLPDDQV GLQVRK DNWITV P H FIVNIGDQIQVLSNA YKSVEHRVIVNS++ER+
Sbjct: 971 LLPDDQVAGLQVRKFDNWITVNPARHGFIVNIGDQIQVLSNATYKSVEHRVIVNSDQERL 1150
Query: 310 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 368
SLAFFYNPR DIPIEP K+L+ ++PALY AMTFDEYR FIRM+GPCGKS VES+KS +
Sbjct: 1151SLAFFYNPRSDIPIEPLKQLITPERPALYPAMTFDEYRLFIRMRGPCGKSQVESMKSSK 1327
>TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin
dioxygenase-like protein {Arabidopsis thaliana}, partial
(82%)
Length = 1326
Score = 504 bits (1299), Expect = e-143
Identities = 239/359 (66%), Positives = 303/359 (83%), Gaps = 3/359 (0%)
Frame = +2
Query: 11 WPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDAN--NIPIIDLRGLLYHGD 68
WPEPIVRVQ+L+E+ + SIP YIKP + RP+ + ++ +D + NIP+IDL H
Sbjct: 116 WPEPIVRVQALAESGLTSIPSCYIKPRSQRPTKTTFATQNDHDHINIPVIDLE----HLS 283
Query: 69 PDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANS 128
+ P R + L+++SEAC+EWGFFQ+VNHG+S +LM+ A+E WR+FF+LP+E+K+++ANS
Sbjct: 284 SEDPVLRETVLKRVSEACREWGFFQVVNHGISHELMESAKEVWREFFNLPLEVKEEFANS 463
Query: 129 PKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVK 188
P TYEGYGSRLGV+KGAILDWSDY+FLH +P SL++ KWPA P S R++ EYG E+VK
Sbjct: 464 PSTYEGYGSRLGVKKGAILDWSDYFFLHSMPPSLRNQAKWPATPSSLRKIIAEYGEEVVK 643
Query: 189 LCGRLMKVLSLNLELEEDFLQNGFGGE-DIGACLRVNFYPKCPKPELTLGLSSHSDPGGM 247
L GR+++++S NL L+ED+L N FGGE ++GACLRVNFYPKCP+P+LTLGLS HSDPGGM
Sbjct: 644 LGGRMLELMSTNLGLKEDYLMNAFGGENELGACLRVNFYPKCPQPDLTLGLSPHSDPGGM 823
Query: 248 TLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKE 307
T+LLPDD V+GLQVRKG++WITVKPVP+AFI+NIGDQIQVLSNAIYKSVEHRVIVNS K+
Sbjct: 824 TILLPDDFVSGLQVRKGNDWITVKPVPNAFIINIGDQIQVLSNAIYKSVEHRVIVNSIKD 1003
Query: 308 RVSLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKS 366
RVSLA FYNP+ D+ IEPAKELV +++PALY+AMT+DEYR FIRMKGPCGK+ VESL S
Sbjct: 1004RVSLAMFYNPKSDLLIEPAKELVTKERPALYSAMTYDEYRLFIRMKGPCGKTQVESLAS 1180
>TC78109 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (48%)
Length = 1314
Score = 179 bits (453), Expect(2) = 2e-66
Identities = 91/255 (35%), Positives = 153/255 (59%)
Frame = +3
Query: 14 PIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPE 73
P+ VQ L++ + +P+RY++P DRP ++S+++S+ +P+IDL LL D + PE
Sbjct: 69 PVPFVQELAKEALTKVPDRYVRPHHDRPILSSTTTSTPLPQLPVIDLSKLLSSHDLNEPE 248
Query: 74 ARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYE 133
L+++ ACKEWGFFQ++NHGVS LM+ ++ +FF+LPME K+++ + E
Sbjct: 249 -----LKKLHYACKEWGFFQLINHGVSESLMENVKKGAEEFFNLPMEEKKKFGQTEGDVE 413
Query: 134 GYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRL 193
GYG V + LDW+D L LP + + + +P R + Y ++ L ++
Sbjct: 414 GYGQAFVVSEEQKLDWADMLVLFTLPPHKRKPHLFSNIPLPFRVDLENYCEKMRTLAIQI 593
Query: 194 MKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPD 253
M +++ +L ++ ++ FG R+N+YP CP+PE +GL+SHSD GG+T+LL
Sbjct: 594 MDLMAHSLAVDPMEIREFFG--QATQSTRMNYYPPCPQPEFVIGLNSHSDGGGLTILLQG 767
Query: 254 DQVTGLQVRKGDNWI 268
+++ GLQ++K WI
Sbjct: 768 NEMDGLQIKKDGLWI 812
Score = 91.7 bits (226), Expect(2) = 2e-66
Identities = 41/106 (38%), Positives = 72/106 (67%)
Frame = +1
Query: 260 QVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRG 319
+++K D VKP+P+AFI+N+GD +++++N I++S+EHR VNS KER S+A FY+P
Sbjct: 787 KLKKMDCGFHVKPLPNAFIINLGDMLEMMTNGIFRSIEHRATVNSEKERFSIASFYSPAF 966
Query: 320 DIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
+ + PA +V + PA++ ++ EY + + CGKS ++S++
Sbjct: 967 NTILSPAPSIVTPNTPAVFKRISAGEYFKGYLAQELCGKSFLDSIR 1104
>TC78110 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (53%)
Length = 1303
Score = 245 bits (626), Expect = 2e-65
Identities = 125/348 (35%), Positives = 211/348 (59%)
Frame = +2
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ + + + +PERY++P DRP I S+++ +P+ID L +
Sbjct: 107 VQEIVKEPLTRVPERYVRPHHDRPII---STTTPLLELPVIDFSKLF----SQDLTIKGL 265
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L ++ ACKEWGFFQ++NHGVS L++ + ++F++LP+E K++++ EGYG
Sbjct: 266 ELDKLHSACKEWGFFQLINHGVSTSLVENVKMGAKEFYNLPIEEKKKFSQKEGDVEGYGQ 445
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
+ + LDW+D +F+ LP ++ + +P LP R+ + Y EL KL +++ +
Sbjct: 446 AFVMSEEQKLDWADMFFMITLPSHMRKPHLFPKLPLPFRDDLETYSAELKKLAIQIIDFM 625
Query: 198 SLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVT 257
+ L+++ ++ FG + R+N+YP CP+PEL +GL+SHSD GG+T+LL +++
Sbjct: 626 ANALKVDAKEIRELFG--EGTQSTRINYYPPCPQPELVIGLNSHSDGGGLTILLQGNEMD 799
Query: 258 GLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNP 317
GLQ++K WI VKP+P+AFI+N+GD +++++N IY S+EHR VN KER+S+A FY+P
Sbjct: 800 GLQIKKDGFWIPVKPLPNAFIINLGDMLEIITNGIYPSIEHRATVNLKKERLSIATFYSP 979
Query: 318 RGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
+ + P+ LV PAL+ + ++ + K GKS ++SL+
Sbjct: 980 SSAVILRPSPTLVTPKTPALFKPIGVTDFYKGYLGKELRGKSFLDSLR 1123
>TC82784 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (59%)
Length = 1206
Score = 241 bits (615), Expect = 3e-64
Identities = 130/349 (37%), Positives = 213/349 (60%), Gaps = 1/349 (0%)
Frame = +3
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ L++ I ++P RYI+ + IN ++S + IP+ID++ LL E +
Sbjct: 27 VQELAKEKISTVPARYIQSQHEELVINEANSILE---IPVIDMKKLL------SLEYGSL 179
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L ++ ACK+WGFFQ+VNH VS L++ + FF+LPM K+++ +P+ EG+G
Sbjct: 180 ELSKLHLACKDWGFFQLVNHDVSSSLLEKVKMEIMDFFNLPMSEKKKFWQTPQHMEGFGQ 359
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
+ + LDW+D +F+ LP L+ + +P LP R+ + Y +E+ L ++ +
Sbjct: 360 AFVLSEEQKLDWADLFFMTTLPKHLRMPHLFPQLPLPLRDTLELYSQEIKNLAMVILGHI 539
Query: 198 SLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLP-DDQV 256
+L++EE ++ F ED +R N+YP CP+PE +GL++HSDP G+T+LL ++
Sbjct: 540 EKSLKMEEMEIRELF--EDGIQMMRTNYYPPCPQPEKVIGLTNHSDPVGLTILLQLNESR 713
Query: 257 TGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYN 316
GLQ+RK W+ VKP+P+AFIVNIGD +++++N IY+S+EHR IVNS KER+S+A FY+
Sbjct: 714 KGLQIRKNCMWVPVKPLPNAFIVNIGDMLEIITNGIYRSIEHRAIVNSEKERLSIATFYS 893
Query: 317 PRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
R + P K L+ E PA + + +EY + + GKS+++ ++
Sbjct: 894 SRHGSILGPVKSLITEQTPARFKKVGVEEYFTNLFARKLEGKSYIDVMR 1040
>TC91229 similar to GP|9294689|dbj|BAB03055.1 gene_id:MHC9.10~similar to
oxidases {Arabidopsis thaliana}, partial (86%)
Length = 1198
Score = 240 bits (613), Expect = 6e-64
Identities = 122/332 (36%), Positives = 193/332 (57%), Gaps = 1/332 (0%)
Frame = +2
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ L T ++P+R+++ +T+RP++ S +S ++P+ID + E +
Sbjct: 65 VQELRRTKPKTVPQRFVRDMTERPTLLSPQNS----DMPVIDFSKF----SKGNKEEVFN 220
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L ++S AC+EWGFFQ++NH + +LM+ + R+FF LP++ KQ+Y +P T +GYG
Sbjct: 221 ELSKLSTACEEWGFFQVINHEIDLNLMENIEDMSREFFMLPLDEKQKYPMAPGTVQGYGQ 400
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
+ LDW + + L P +++ N WP P E + Y R++ KLC L+K +
Sbjct: 401 AFVFSEDQKLDWCNMFALGIEPHYIRNSNLWPKKPARLSETIELYSRKIRKLCQNLLKYI 580
Query: 198 SLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLL-LPDDQV 256
+L L LEED + FG + +R+N+YP C +P+L LGLS HSD +T+L
Sbjct: 581 ALGLSLEEDVFEKMFG--EAVQAIRMNYYPTCSRPDLVLGLSPHSDGSALTVLQQAKGSP 754
Query: 257 TGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYN 316
GLQ+ K + W+ V+P+ +A ++NIGD I+VL+N YKSVEHR + + K+R+S+ FY
Sbjct: 755 VGLQILKDNTWVPVEPISNALVINIGDTIEVLTNGKYKSVEHRAVAHEEKDRLSIVTFYA 934
Query: 317 PRGDIPIEPAKELVKEDKPALYTAMTFDEYRR 348
P D+ + P ELV E+ P Y EY +
Sbjct: 935 PSYDLELGPMLELVDENHPCKYKRYNHGEYSK 1030
>TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1323
Score = 239 bits (610), Expect = 1e-63
Identities = 122/348 (35%), Positives = 213/348 (61%)
Frame = +3
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ L + I +PERY++ D + SS+ SS +PIIDL LL D +
Sbjct: 177 VQELVKQPITKVPERYLQQNQDPSLVVSSTISSQ--QVPIIDLNKLLSEED-------GT 329
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L++ ACKEWGFFQ++NHGV L++ ++ ++FF+L E K+ + P EG+G
Sbjct: 330 ELQKFDLACKEWGFFQLINHGVDLSLVENFKKHVQEFFNLSAEEKKIFGQKPGDMEGFGQ 509
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
V + L+W+D +++ LP +++ N +P++P R+ + Y E+ KLC +++ +
Sbjct: 510 MFVVSEEHKLEWADLFYIVTLPTYIRNPNLFPSIPQPFRDNLEMYAIEIKKLCVTIVEFM 689
Query: 198 SLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVT 257
+ L+++ + L + F E+ +R+N+YP CP+PE +GL+ HSD G +T+LL +++
Sbjct: 690 AKALKIQPNELLDFF--EEGSQAMRMNYYPPCPQPEQVIGLNPHSDVGALTILLQLNEIE 863
Query: 258 GLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNP 317
GLQVRK WI +KP+ +AF++NIGD +++++N IY+S+EHR +N+ KER+S+A F +
Sbjct: 864 GLQVRKDGMWIPIKPLSNAFVINIGDMLEIMTNGIYRSIEHRATINAEKERISIATFNSG 1043
Query: 318 RGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
R + + PA LV + PA++ + +++ + + GKSH++ ++
Sbjct: 1044RLNAILCPAPSLVTPESPAVFNNVGVEDFFKAYFSRKLEGKSHIDVMR 1187
>TC80530 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis
thaliana, partial (52%)
Length = 1219
Score = 237 bits (604), Expect = 6e-63
Identities = 117/334 (35%), Positives = 203/334 (60%), Gaps = 5/334 (1%)
Frame = +2
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ L+E I +PE+Y++P D +++++SS +PIID LL +
Sbjct: 32 VQKLAEQGITKVPEQYLQPNQDSILVSNTTSSPQ---LPIIDFDKLLCEDGIE------- 181
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L ++ ACKEWGFFQ++NHGV+P L++ + ++FFHLPME K+++ + + +G+G
Sbjct: 182 -LEKLDNACKEWGFFQLINHGVNPSLVESVKIGVQQFFHLPMEEKKKFWQTEEELQGFGQ 358
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
+ L W D +++ PL ++ + P +P R+ F+ Y EL KLC ++++ +
Sbjct: 359 VYVALEEQKLRWGDMFYVKTFPLHIRLPHLIPCMPQPFRDDFENYSLELKKLCFKIIERM 538
Query: 198 SLNLELEE-----DFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLP 252
+ L++++ DF + G +R+N+YP CP+P+ +GL+ HSD +T+LL
Sbjct: 539 TKALKIQQPNELLDFFEEG------DQSIRMNYYPPCPQPDQVIGLNPHSDASALTILLQ 700
Query: 253 DDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLA 312
+++ GLQ++K W+ + P+P+AF+VNIGD +++++N IY+S+EHR NS KER+S+A
Sbjct: 701 VNEMQGLQIKKDGMWVPINPLPNAFVVNIGDLLEIMTNGIYRSIEHRATANSEKERISVA 880
Query: 313 FFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEY 346
F+N + + PA LV + PA++ +T +EY
Sbjct: 881 GFHNIQMGRDLGPAPSLVTPETPAMFKTITLEEY 982
>TC77355 weakly similar to PIR|T05552|T05552 SRG1 protein-related protein
F24A6.150 - Arabidopsis thaliana, partial (44%)
Length = 1359
Score = 236 bits (601), Expect = 1e-62
Identities = 127/352 (36%), Positives = 187/352 (53%)
Frame = +1
Query: 14 PIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPE 73
P+ VQ + + +P +Y++ + +N S + IP+ID L
Sbjct: 82 PVPNVQEMVKVNPLQVPTKYVRNEEEMEKVNYMPQLS--SEIPVIDFTQL--------SN 231
Query: 74 ARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYE 133
L ++ ACKEWGFFQ+VNHGV DL+ R+ +FF LPME K++YA P +
Sbjct: 232 GNMEELLKLEIACKEWGFFQMVNHGVEKDLIQRMRDASNEFFELPMEEKEKYAMLPNDIQ 411
Query: 134 GYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRL 193
GYG V + LDWSD L P + WP +E+ + Y E+ ++ L
Sbjct: 412 GYGQNYVVSEEQTLDWSDVLVLIIYPDRYRKLQFWPKTSHGFKEIIEAYSSEVRRVGEEL 591
Query: 194 MKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPD 253
+ LS+ + LE+ L +++ LRVN+YP C PE LGLS HSD +TLL+ D
Sbjct: 592 LSFLSIIMGLEKHALAELH--KELIQGLRVNYYPPCNNPEQVLGLSPHSDTTTITLLIQD 765
Query: 254 DQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAF 313
D V GL++R NW+ VKP+ A ++N+GD I++LSN YKSVEHRV+ N NK R+S A
Sbjct: 766 DDVLGLEIRNKGNWVPVKPISDALVINVGDVIEILSNGKYKSVEHRVMTNQNKRRISYAS 945
Query: 314 FYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
F PR D+ +EP ++ P +Y + + +Y R + K+H + K
Sbjct: 946 FLFPRDDVEVEPFDHMIDAQNPKMYQKVKYGDYLRHSLKRKMERKTHTDVAK 1101
>TC86474 weakly similar to GP|6016680|gb|AAF01507.1| putative
leucoanthocyanidin dioxygenase {Arabidopsis thaliana},
partial (20%)
Length = 1395
Score = 234 bits (596), Expect = 5e-62
Identities = 130/355 (36%), Positives = 192/355 (53%), Gaps = 3/355 (0%)
Frame = +1
Query: 14 PIVRVQSLSETCIDS---IPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPD 70
P + V ++ ET + +PERY++ + + S + +P+ID GLL HG+ +
Sbjct: 109 PCLPVPNIQETVRKNPLKVPERYVRSEEEIEKVLYMPHFS--SQVPVIDF-GLLSHGNKN 279
Query: 71 HPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK 130
L ++ ACKEWGFFQIVNHG+ DLM ++ +FF L +E K +YA P
Sbjct: 280 E-------LLKLDIACKEWGFFQIVNHGMEIDLMQRLKDVVAEFFDLSIEEKDKYAMPPD 438
Query: 131 TYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLC 190
+GYG V + ILDW D L P + WP P ++ + Y E+ ++
Sbjct: 439 DIQGYGHTSVVSEKQILDWCDQLILLVYPTRFRKPQFWPETPEKLKDTIEAYSSEIKRVG 618
Query: 191 GRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLL 250
L+ LSL LEE L +++ LRVN+YP C PE +GL+ HSD +T++
Sbjct: 619 EELINSLSLIFGLEEHVLLGLH--KEVLQGLRVNYYPPCNTPEQVIGLTPHSDASTVTIV 792
Query: 251 LPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVS 310
+ DD VTGL+VR NW+ + P+P+A +VN+GD I+VLSN YKSVEHR + N NK R S
Sbjct: 793 MQDDDVTGLEVRYKGNWVPINPIPNALVVNLGDVIEVLSNGKYKSVEHRAMTNKNKRRTS 972
Query: 311 LAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
F PR D + P ++ + P +Y +T+ EY R + GK+ ++ K
Sbjct: 973 FVSFLFPRDDAELGPFDHMIDDQNPKMYKEITYGEYLRHTLNRKLEGKTQTDATK 1137
>TC78010 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1321
Score = 231 bits (589), Expect = 3e-61
Identities = 116/329 (35%), Positives = 204/329 (61%)
Frame = +3
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ L + I +PERY++ D PS+ SS+ S +P+IDL LL D +
Sbjct: 114 VQELVKQPITKVPERYLQQNQD-PSLVVSSTKS-LPQVPVIDLSKLLSEEDE-------T 266
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L+++ ACKEWGFFQ++NHGV+P L++ ++ + FF+LP+E K+ + P EG+G
Sbjct: 267 ELQKLDHACKEWGFFQLINHGVNPLLVENFKKLVQDFFNLPVEEKKILSQKPGNIEGFGQ 446
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
V + L+W+D + + P +++ +P++P RE + Y L KLC +++ +
Sbjct: 447 LFVVSEDHKLEWADLFHIITHPSYMRNPQLFPSIPQPFRESLEMYSLVLKKLCVMIIEFM 626
Query: 198 SLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVT 257
S L+++++ L F E+ G +R+N+YP CP+P+ +GL+ HSD +T+LL +++
Sbjct: 627 SKALKIQKNELLEFF--EEGGQSMRMNYYPPCPQPDKVIGLNPHSDGTALTILLQLNEIE 800
Query: 258 GLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNP 317
GLQ++K WI +KP+ +AF++NIGD +++++N IY+S+EHR +NS KER+S+A F++
Sbjct: 801 GLQIKKDGMWIPIKPLTNAFVINIGDMLEIMTNGIYRSIEHRATINSEKERISIATFHSA 980
Query: 318 RGDIPIEPAKELVKEDKPALYTAMTFDEY 346
R + + P L+ PA++ ++ +++
Sbjct: 981 RLNAILAPVPSLITPKTPAVFNDISVEDF 1067
>TC85652 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (50%)
Length = 1489
Score = 229 bits (584), Expect = 1e-60
Identities = 118/348 (33%), Positives = 203/348 (57%)
Frame = +1
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
V L++ I +PE++++P D + ++ S +P+IDL LL A+
Sbjct: 298 VHELAKQSIVEVPEQFLRPNQDATHVINTDSLPQ---VPVIDLGKLLGED--------AT 444
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L ++ ACKEWGFFQI+NHGV+ L++ + ++F LP+E K+++ +P EG+G
Sbjct: 445 ELEKLDLACKEWGFFQIINHGVNTSLIEKVKIGIKEFLSLPVEEKKKFWQTPNDMEGFGQ 624
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
V L+W+D + + LPL ++ +P++ R+ + Y E+ KLC ++ +
Sbjct: 625 MFVVSDDQKLEWADLFLITTLPLDERNPRLFPSIFQPFRDNLEIYCSEVQKLCFTIISQM 804
Query: 198 SLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVT 257
L++E + + F I +R N YP CP+PE +GL+ HSD G +T+LL +++
Sbjct: 805 EKALKIESNEVTELFN--HITQAMRWNLYPPCPQPENVIGLNPHSDVGALTILLQANEIE 978
Query: 258 GLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNP 317
GLQ+RK WI V+P+P+AF++NIGD +++++N IY+S+EHR VNS KER+S+A F+ P
Sbjct: 979 GLQIRKDGQWIPVQPLPNAFVINIGDMLEIVTNGIYRSIEHRATVNSEKERISIAAFHRP 1158
Query: 318 RGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
+ + P LV ++PA + ++ EY + + GKS ++ ++
Sbjct: 1159HVNTILSPRPSLVTPERPASFKSIAVGEYLKAYFSRKLEGKSCLDVMR 1302
>TC80257 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (50%)
Length = 1376
Score = 154 bits (388), Expect(2) = 1e-58
Identities = 79/195 (40%), Positives = 121/195 (61%), Gaps = 1/195 (0%)
Frame = +2
Query: 172 PPSCREVFDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGGEDIGA-CLRVNFYPKCP 230
P RE + Y EL KL +++++++ L + + F +IG +RVN+YP CP
Sbjct: 596 PHHFRENLEVYSVELEKLAIKVIELMANALAINPKEMTELF---NIGTQMVRVNYYPPCP 766
Query: 231 KPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSN 290
+PE +GL SHSD GG+T+LL + GLQ+RK WI VKP+P+AFIVNIGD +++++N
Sbjct: 767 QPERVIGLKSHSDAGGLTILLQTSDIDGLQIRKDGQWIPVKPLPNAFIVNIGDMLEIITN 946
Query: 291 AIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFI 350
IY S+EHR VNS KER+S+A FY P + PA LV +++PA + ++ ++
Sbjct: 947 GIYPSIEHRATVNSEKERISVAAFYGPNMQAMLAPAPSLVTQERPAQFRRISVVDHFNGY 1126
Query: 351 RMKGPCGKSHVESLK 365
+ GKS++ +K
Sbjct: 1127FSQELRGKSYLNEMK 1171
Score = 90.9 bits (224), Expect(2) = 1e-58
Identities = 52/162 (32%), Positives = 86/162 (52%)
Frame = +1
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ L++ + +PERY+ P + +++++SS +P+IDL LL +
Sbjct: 154 VQELAKEQLTKVPERYVLPDHETVVLSNTTSSPQ---VPVIDLAELL----SQDINLKRH 312
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L ++ ACKEWGFFQ+VNHG+S L++ ++ + F L ME K+ EG+G
Sbjct: 313 ELEKLHYACKEWGFFQLVNHGISTSLVEDMKKGAKTLFELSMEEKKNLWQIEGEMEGFGQ 492
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVF 179
+ + L+W+D +FL LP L+ + +PPS + F
Sbjct: 493 SFVLSEEQKLEWADTFFLSTLPSHLRKPYLFNQIPPSFQRKF 618
>TC91081 similar to GP|5813796|gb|AAD52015.1| unknown {Pisum sativum},
partial (93%)
Length = 886
Score = 215 bits (547), Expect(2) = 2e-58
Identities = 105/119 (88%), Positives = 111/119 (93%)
Frame = +1
Query: 250 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 309
LLPDDQ+ GLQVRKGDNWITVKPV +AFIVNIGDQIQVLSNAIY+SVEHRVI NS+KERV
Sbjct: 37 LLPDDQIHGLQVRKGDNWITVKPVRNAFIVNIGDQIQVLSNAIYRSVEHRVIANSHKERV 216
Query: 310 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 368
SLAFFYNP+ DIPIEPAKELVK DKPALY AMTFDEYR FIRM+GPCGKS VESLKSPR
Sbjct: 217 SLAFFYNPKSDIPIEPAKELVKPDKPALYPAMTFDEYRLFIRMRGPCGKSQVESLKSPR 393
Score = 28.9 bits (63), Expect(2) = 2e-58
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = +2
Query: 238 LSSHSDPGGMTL 249
LSSHSDPGGMTL
Sbjct: 2 LSSHSDPGGMTL 37
>TC85651 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (39%)
Length = 1269
Score = 221 bits (562), Expect = 5e-58
Identities = 118/348 (33%), Positives = 202/348 (57%)
Frame = +3
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ L + I +P++Y++P D +++++S +P+IDL LL A
Sbjct: 57 VQELEKQGITKVPKQYLQPNQDSILVSNTTSLPQ---LPVIDLSKLLCED--------AI 203
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L + ACK+WGFFQ++NHGV+P L++ + +KFF+LP+E K+++ + + +G+G
Sbjct: 204 ELENLDHACKDWGFFQLINHGVNPLLVENIKIGVQKFFNLPIEEKKKFWQTTEEMQGFGQ 383
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
+ L W D +F+ PL + + P +P R+ + Y E+ KLC L++ +
Sbjct: 384 VYVALEEEKLRWGDMFFIKTFPLHTRHPHLIPCIPQPFRDNLESYSLEVNKLCVTLIEFM 563
Query: 198 SLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVT 257
S L+++ + L + F E+ +R+N+YP CP+P+ +GL+ HSD G +T+LL +++
Sbjct: 564 SKALKIKPNELLDIF--EEGSQAMRMNYYPPCPQPDQVIGLNPHSDAGTLTILLQVNEME 737
Query: 258 GLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNP 317
GLQ++K WI ++P+ AF+VN+GD ++V +N IY+S+EHR VNS KER+S+A F+ P
Sbjct: 738 GLQIKKDGIWIPIRPLSDAFVVNVGDILEVQTNGIYRSIEHRATVNSEKERISVAAFHAP 917
Query: 318 RGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
I P LV PAL+ M ++ GKS+++ ++
Sbjct: 918 HMGGYIGPTPSLVTPQSPALFKTMPTADFLNGYIASRIKGKSYLDVVR 1061
>TC85654 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (32%)
Length = 1307
Score = 140 bits (354), Expect(2) = 5e-58
Identities = 62/145 (42%), Positives = 101/145 (68%)
Frame = +1
Query: 221 LRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVN 280
+R+N+YP CP+PE +G+S HSD G +T+LL + V GLQ+RK WI+V+P+P+AF++N
Sbjct: 685 MRINYYPPCPQPEHVIGVSPHSDMGALTILLQANDVEGLQIRKDGQWISVQPLPNAFVIN 864
Query: 281 IGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPALYTA 340
IGD +++L+N IY+S+EHR VNS KER+S+A F+ + I P L+ ++PAL+
Sbjct: 865 IGDMLEILTNGIYRSIEHRGTVNSKKERISIATFHRLQMSSVIGPTPSLITAERPALFKT 1044
Query: 341 MTFDEYRRFIRMKGPCGKSHVESLK 365
++ +Y + GKS+++++K
Sbjct: 1045ISVADYINRYLSRQLDGKSNLDNVK 1119
Score = 101 bits (252), Expect(2) = 5e-58
Identities = 60/205 (29%), Positives = 108/205 (52%), Gaps = 6/205 (2%)
Frame = +3
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
VQ L + I +PE+Y++P D + +++S +P+I+L LL + +
Sbjct: 111 VQELVKQPITKVPEQYLQPNQDLVVVCNTTSLP---KVPVINLHKLLSNDTIE------- 260
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGS 137
L AC+EWGFFQ++NHGV+ L++ ++ +FF+LP++ K++Y +P +G+G
Sbjct: 261 -LENFDHACREWGFFQLINHGVNTLLVENMKKGVEQFFNLPIDEKKKYWQTPNDMQGFGQ 437
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLC----GRL 193
V L+W D ++++ LPL + + P++P R+ + Y EL +L GR+
Sbjct: 438 LFVVSDEQKLEWQDMFYINTLPLDSRHPHLIPSIPKPFRDHLETYCLELKQLAFTIIGRM 617
Query: 194 MKVLSLNLELEEDFLQNGF--GGED 216
K L + +F ++ G ED
Sbjct: 618 EKTLKIKTNELVEFFEDAIHQGNED 692
>TC91042 similar to PIR|T49209|T49209 leucoanthocyanidin dioxygenase-like
protein - Arabidopsis thaliana, partial (35%)
Length = 732
Score = 209 bits (533), Expect = 1e-54
Identities = 123/209 (58%), Positives = 141/209 (66%), Gaps = 11/209 (5%)
Frame = +1
Query: 1 MMKKNISPLNWPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDL 60
++KK ISP +WPEPI+RVQSLSE+C DSIP+RYIKPL+DRPSINS + NNIPIIDL
Sbjct: 151 LLKKMISPQDWPEPIIRVQSLSESCKDSIPQRYIKPLSDRPSINSILET--INNIPIIDL 324
Query: 61 RGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPME 120
RGL +P +AST +QISEACKEWGFFQIVNHGVS DLMD+A+ETWR+FFHLPME
Sbjct: 325 RGLY----STNPHEKASTFKQISEACKEWGFFQIVNHGVSHDLMDLAKETWRQFFHLPME 492
Query: 121 LKQQYANSPKTYEGYGSRLGVEKGAILDW---SDYYFLHYLPLSLKDYNKWPALPPSC-- 175
+KQ YANSPKTYEGYGSR W Y L L LS + L
Sbjct: 493 VKQLYANSPKTYEGYGSR---------PWC*ERCYS*LE*LLLSSLSSFVFEGLQQMAFS 645
Query: 176 -----REVFDEYGRELVK-LCGRLMKVLS 198
VFDEYG EL + + LMKVLS
Sbjct: 646 ASLLQGRVFDEYGIELGEGMWSGLMKVLS 732
>TC85507 homologue to SP|P31239|ACCO_PEA 1-aminocyclopropane-1-carboxylate
oxidase (EC 1.-.-.-) (ACC oxidase) (Ethylene-forming
enzyme) (EFE), complete
Length = 1342
Score = 209 bits (533), Expect = 1e-54
Identities = 117/314 (37%), Positives = 180/314 (57%), Gaps = 3/314 (0%)
Frame = +1
Query: 38 TDRPSINSSS-SSSDANNIPIIDLRGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVN 96
++R N S S N P++D+ L + E R +T+ I +AC+ WGFF+ VN
Sbjct: 52 SERKERNKES*KRSKMENFPVVDMGKL-------NTEERKATMEMIKDACENWGFFECVN 210
Query: 97 HGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGSRLGVEKGAILDWSDYYFLH 156
H +S +LMD + ++ + ME + + + K E S + LDW +FL
Sbjct: 211 HSISIELMDKVEKLTKEHYKKCMEQRFKEMVASKGLECVQSEIND-----LDWESTFFLR 375
Query: 157 YLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGF-GGE 215
+LP S + ++ P L R+ E+ +L KL L+ +L NL LE+ +L+ F G +
Sbjct: 376 HLPSS--NISEIPDLDEDYRKTMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSK 549
Query: 216 DIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPH 275
+V+ YP CPKP+L GL +H+D GG+ LL DD+V+GLQ+ K D WI V P+PH
Sbjct: 550 GPNFGTKVSNYPPCPKPDLIKGLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPH 729
Query: 276 AFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKP 335
+ ++N+GDQ++V++N YKSV HRVI ++ R+S+A FYNP D I PA L+KED+
Sbjct: 730 SIVINLGDQLEVITNGKYKSVMHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDET 909
Query: 336 A-LYTAMTFDEYRR 348
+ +Y FD+Y +
Sbjct: 910 SEIYPKFIFDDYMK 951
>TC91108 similar to PIR|T05552|T05552 SRG1 protein-related protein F24A6.150
- Arabidopsis thaliana, partial (14%)
Length = 1133
Score = 207 bits (526), Expect = 7e-54
Identities = 116/334 (34%), Positives = 179/334 (52%)
Frame = +2
Query: 28 SIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARASTLRQISEACK 87
++PE Y++ T+ + ++++PIID GLL +G+ + L ++ ACK
Sbjct: 194 NVPESYVR--TEEQMEKHLYTPHLSSHLPIIDF-GLLSNGNKEE-------LLKLDFACK 343
Query: 88 EWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGSRLGVEKGAIL 147
EWG+FQIVNHG+ DLM ++ +F L ++ K +YA P +GYG +
Sbjct: 344 EWGYFQIVNHGMQIDLMQRVKDAVAEFSELSIKEKIKYAMPPDDMQGYGH-------TSV 502
Query: 148 DWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELEEDF 207
DWSDY L P + WP ++ D Y E+ ++ ++ LSL L LEE
Sbjct: 503 DWSDYLVLLVYPTRFRKPQFWPK---ELKDTIDAYSNEIKRIGEEVINSLSLLLGLEEHG 673
Query: 208 LQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNW 267
L + ++ RVN+YP C PE LG+S HSDP + +++ DD V+G ++R NW
Sbjct: 674 LLDLH--REVHQGFRVNYYPPCNTPEKVLGVSPHSDPRTIAIVMQDDDVSGTEIRYKRNW 847
Query: 268 ITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAK 327
+ + P+P A +VN+GD I++LSN YKSVEHR I N NK+R + + P+GD I
Sbjct: 848 VPITPIPGALVVNVGDVIEILSNGKYKSVEHRAITNKNKKRTTFISYLFPQGDAEIGALD 1027
Query: 328 ELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHV 361
++ + P +Y T+ EY R + GK H+
Sbjct: 1028HMIDDQNPKMYKDTTYGEYLRHVLNTKLEGKPHI 1129
>TC89553 weakly similar to SP|P24397|HY6H_HYONI Hyoscyamine 6-dioxygenase
(EC 1.14.11.11) (Hyoscyamine 6-beta- hydroxylase).
[Henbane], partial (49%)
Length = 1119
Score = 206 bits (524), Expect = 1e-53
Identities = 126/331 (38%), Positives = 185/331 (55%), Gaps = 3/331 (0%)
Frame = +1
Query: 28 SIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARASTLRQISEACK 87
S+P +++PL RP ++ SS IP+IDL G DH A T+ QI +A +
Sbjct: 58 SVPPSFVQPLECRPGKVTNPSSK---TIPLIDL------GGHDH----AHTILQILKASE 198
Query: 88 EWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELK-QQYANSPK--TYEGYGSRLGVEKG 144
E+GFFQ++NHGVS DLMD A +++F +P + K + + P + Y S +
Sbjct: 199 EYGFFQVINHGVSKDLMDEAFNIFQEFHAMPPKEKISECSRDPNGINCKIYASSENYKID 378
Query: 145 AILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELE 204
A+ W D H P S + WP PP REV +Y +EL KL ++++L L L+
Sbjct: 379 AVQYWKDT-LTHPCPPSGEFMEFWPQKPPKYREVVGKYTQELNKLGHEILEMLCEGLGLK 555
Query: 205 EDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKG 264
++ G + + +P CP+P LTLGL+ H DP +T+LL D +V GLQ+ K
Sbjct: 556 LEYF---IGELSENPVILGHHFPPCPEPSLTLGLAKHRDPTIITILLQDQEVHGLQILKD 726
Query: 265 DNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIE 324
D WI V+P+P+A +VNIG +Q+++N EHRV+ NS R S+A+F P IE
Sbjct: 727 DEWIPVEPIPNALVVNIGLILQIITNGRLVGAEHRVVTNSKSVRTSVAYFIYPSFSRMIE 906
Query: 325 PAKELVKEDKPALYTAMTFDEYRRFIRMKGP 355
PAKELV + P +Y +M+F E+R+ KGP
Sbjct: 907 PAKELVDGNNPPIYKSMSFGEFRKKFYDKGP 999
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,883,177
Number of Sequences: 36976
Number of extensions: 201167
Number of successful extensions: 1249
Number of sequences better than 10.0: 180
Number of HSP's better than 10.0 without gapping: 1072
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1085
length of query: 368
length of database: 9,014,727
effective HSP length: 97
effective length of query: 271
effective length of database: 5,428,055
effective search space: 1471002905
effective search space used: 1471002905
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0204.4


