

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0204.2
(664 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
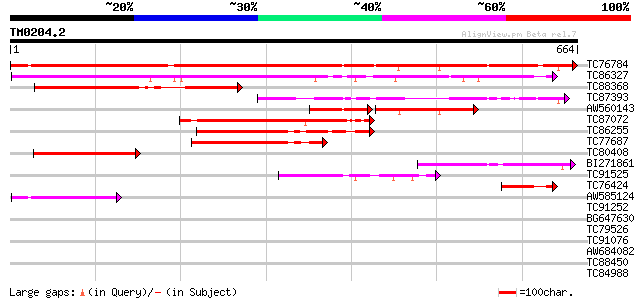
Score E
Sequences producing significant alignments: (bits) Value
TC76784 similar to PIR|G84825|G84825 probable CCCH-type zinc fin... 1047 0.0
TC86327 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20 ... 468 e-132
TC88368 similar to GP|15810487|gb|AAL07131.1 unknown protein {Ar... 276 1e-74
TC87393 similar to PIR|G84825|G84825 probable CCCH-type zinc fin... 253 2e-67
AW560143 similar to PIR|G84825|G848 probable CCCH-type zinc fing... 128 2e-60
TC87072 similar to PIR|D84581|D84581 probable CCCH-type zinc fin... 199 3e-51
TC86255 similar to PIR|D84581|D84581 probable CCCH-type zinc fin... 196 2e-50
TC77687 similar to PIR|D84581|D84581 probable CCCH-type zinc fin... 173 2e-43
TC80408 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20 ... 136 2e-32
BI271861 130 1e-30
TC91525 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20 ... 106 3e-23
TC76424 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20 ... 65 8e-11
AW585124 GP|13487732|gb ORF1 {TT virus}, partial (2%) 57 2e-08
TC91252 similar to GP|9988428|dbj|BAB12694.1 putative zinc finge... 40 0.002
BG647630 similar to GP|20521270|dbj hypothetical protein~similar... 40 0.004
TC79526 weakly similar to GP|14586362|emb|CAC42893. putative pro... 38 0.011
TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [... 37 0.024
AW684082 homologue to PIR|T47773|T47 hypothetical protein F24I3.... 36 0.042
TC88450 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 36 0.054
TC84988 similar to GP|14018368|emb|CAC38358. zinc finger protein... 35 0.12
>TC76784 similar to PIR|G84825|G84825 probable CCCH-type zinc finger protein
[imported] - Arabidopsis thaliana, partial (56%)
Length = 2631
Score = 1047 bits (2708), Expect = 0.0
Identities = 550/699 (78%), Positives = 594/699 (84%), Gaps = 35/699 (5%)
Frame = +3
Query: 1 MCSGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGF 60
MCSGSKSKLSSS+ ME + + G+HN SILLELSAS+DF+A KREVDEK LDVNE GF
Sbjct: 216 MCSGSKSKLSSSDCAMEDKLQM--GIHNSSILLELSASDDFDAFKREVDEKDLDVNEEGF 389
Query: 61 WYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAG 120
WYGRRIGSKKM SEKRTPLMIAS+FGSTRVV+Y+V G+VDVN CGSD ATALHCAVAG
Sbjct: 390 WYGRRIGSKKMESEKRTPLMIASMFGSTRVVEYIVSAGKVDVNGVCGSDMATALHCAVAG 569
Query: 121 GSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESGQL 180
GSE LEVVK+LLDAG+DAD LDA GNKP++LIAPAFNSSSKSRRK +E+ LRG S +L
Sbjct: 570 GSEFLLEVVKLLLDAGADADCLDASGNKPVDLIAPAFNSSSKSRRKVLEMFLRGEVSAEL 749
Query: 181 IHQEMDLELISAPFS-SKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYS 239
I EMD APFS KEG DKKE+P+D+SLPDINNG+YGSDEFRMYSFKVKPCSRAYS
Sbjct: 750 IQGEMD-----APFSLKKEGGDKKEFPIDISLPDINNGVYGSDEFRMYSFKVKPCSRAYS 914
Query: 240 HDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQY 299
HDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKG+CQKGDSCEYAHGVFESWLHPAQY
Sbjct: 915 HDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGSCQKGDSCEYAHGVFESWLHPAQY 1094
Query: 300 RTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSS 359
RTRLCKDETGC RKVCFFAHRPEELRPVYASTGSAMPSPKSYS++ +DM+ MSPL+LSSS
Sbjct: 1095RTRLCKDETGCNRKVCFFAHRPEELRPVYASTGSAMPSPKSYSASGMDMTSMSPLSLSSS 1274
Query: 360 SLPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEM 419
SLPMSTVSTPPMSPLAGS SPKSGN+WQNK LNLTPPSLQLPGSRLK+ALSARD DLEM
Sbjct: 1275SLPMSTVSTPPMSPLAGSSSPKSGNMWQNK--LNLTPPSLQLPGSRLKSALSARDLDLEM 1448
Query: 420 EMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFR----NRIGDLTPTNLDDLLGSVDSNL 475
E+LGL SP R+QQQQQQQQQLIEEIARISSPSFR NRI DL PTNLDDLL S D NL
Sbjct: 1449ELLGLDSP-RRQQQQQQQQQLIEEIARISSPSFRNSEFNRIADLNPTNLDDLLASADPNL 1625
Query: 476 LSQLHGLS-GPSTPT-HLQSQSRHQMMQ-----QNMNHLRSSYPS-NIPSSPVRKPSSLG 527
LSQLHGLS PSTPT + S S QM Q QNMNHLR+SYPS N+PSSPVRKPS G
Sbjct: 1626LSQLHGLSMQPSTPTQQMHSPSAMQMRQNMNMGQNMNHLRASYPSNNMPSSPVRKPSPYG 1805
Query: 528 FDSSSAVAAAVMNSRSAAFAK-RSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSP 586
FDSS+AVAAAVMNSRSAAFAK RSQSFIDRGAA THHLG+S PSNPSCRVSSG SDW SP
Sbjct: 1806FDSSAAVAAAVMNSRSAAFAKQRSQSFIDRGAA-THHLGLSPPSNPSCRVSSGLSDWGSP 1982
Query: 587 SGKLDWGVNGDELSKLRKSASFGFRNNG-MAPTGSPMAHSEHAEPDVSWVHSLVKD---- 641
+GKLDWGVNGDEL+KLRKSASFGFRNNG APT S+HAEPDVSWV+SLVKD
Sbjct: 1983TGKLDWGVNGDELNKLRKSASFGFRNNGPAAPTA-----SQHAEPDVSWVNSLVKDVPSD 2147
Query: 642 ----------------DLSKEMLPPWVEQLYIEQEQMVA 664
DLS+++LPPWVEQ+Y EQEQMVA
Sbjct: 2148NSGVYGAENMRQLQQYDLSRDVLPPWVEQMYKEQEQMVA 2264
>TC86327 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20
{Arabidopsis thaliana}, partial (50%)
Length = 3018
Score = 468 bits (1204), Expect = e-132
Identities = 287/699 (41%), Positives = 390/699 (55%), Gaps = 60/699 (8%)
Frame = +2
Query: 3 SGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGFWY 62
S S S S V + + +FS LLE +++NDFE K +D +NE GFWY
Sbjct: 296 SSSSSAAEGSNTVEDMKNLTVRTDDSFSSLLEHASNNDFEDFKVALDSDASLINEVGFWY 475
Query: 63 GRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGS 122
R+ GS ++ E RTPLM+A+ +GS ++K ++ DVN +CG+DK+TALHCA + GS
Sbjct: 476 VRQKGSNQIVLEHRTPLMVAASYGSIDILKLILSYPEADVNFSCGTDKSTALHCAASSGS 655
Query: 123 ELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKS---RRKAMELLLRGGESGQ 179
+++ +K+LL AG+D +S+DA G +P+++I K + ELL G
Sbjct: 656 VNAVDAIKLLLSAGADINSVDANGKRPVDVIVVPIVVPHKLEGVKTILEELLSDSASEGS 835
Query: 180 LIHQEMDLELIS------APFSSKEG-------------------SDKKEYPVDVSLPDI 214
+ + L LIS AP SS E S+KKEYPVD SLPDI
Sbjct: 836 VDDCSLPLSLISSSPGSSAPLSSAENGSPSSPVAPKFTDTAVNSTSEKKEYPVDPSLPDI 1015
Query: 215 NNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRK 274
N +Y +DEFRMYSFKV+PCSRAYSHDWTECPFVHPGENARRRDPRK+ YSCVPCP+FRK
Sbjct: 1016KNSMYATDEFRMYSFKVRPCSRAYSHDWTECPFVHPGENARRRDPRKFHYSCVPCPDFRK 1195
Query: 275 GACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSA 334
GAC++ D CEYAHGVFE WLHPAQYRTRLCKD GC R+VCFFAH PEELRP+Y STGSA
Sbjct: 1196GACRRSDMCEYAHGVFECWLHPAQYRTRLCKDGMGCNRRVCFFAHSPEELRPLYVSTGSA 1375
Query: 335 MPSPKSYSSNAVDMSVMSPLAL------SSSSLPMSTVSTPPMSPLAGSLSPKSGNLWQN 388
+PSP+S +S A M + + ++L S S + S + PP+SP S N W
Sbjct: 1376VPSPRSAASTANVMDMAAAMSLFPGSPSSISLMSQSPFAQPPLSP-----SANGNNAWPQ 1540
Query: 389 KINLNLTPPSLQLPG-----SRLKTALSARDFDLEMEMLGLGSPTRQQQQQQQQQQLIEE 443
P+L LPG SRL+++LSARD + Q QQQ++ +
Sbjct: 1541P-----NVPALHLPGSINQTSRLRSSLSARDMPHD-------DFNNMLQDFDGQQQILND 1684
Query: 444 IARISSP-------SFRNRIGDLTPTNLDDLL-GSVDSNLLSQLHGLSGPSTPTHLQSQ- 494
++ S P R LTP+NLDDL + S+ + +PTH +
Sbjct: 1685LSCFSQPRPGAISVGRSGRPKTLTPSNLDDLFCAEIASSPRYSDPAAASVFSPTHKSAVF 1864
Query: 495 SRHQMMQQNMNHLRSSY--PSNIPSSPVRKPSSLGFDS------SSAVAAAVMNSRSAAF 546
++ Q +Q +++ + ++ P+N+ P+ +S G S S A + M+SR +AF
Sbjct: 1865NQFQQLQSSLSPINTNVMSPTNV-EHPLFHQASYGLSSPGRMSPRSMEALSPMSSRLSAF 2041
Query: 547 A---KRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDW-NSPSGKLDWGVNGDELSKL 602
A K+ Q + + LG ++P + + +S W +SP GK DW VN ++ +
Sbjct: 2042AQREKQQQQQQQLRSLSSRELGANNPLSAVGSPVNSWSKWGSSPIGKADWSVNPNDFGQT 2221
Query: 603 RKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKD 641
++S SF NNG EPDV WVHSLVKD
Sbjct: 2222QRSTSFEHGNNG-------------EEPDVGWVHSLVKD 2299
>TC88368 similar to GP|15810487|gb|AAL07131.1 unknown protein {Arabidopsis
thaliana}, partial (25%)
Length = 665
Score = 276 bits (707), Expect = 1e-74
Identities = 145/243 (59%), Positives = 170/243 (69%)
Frame = +3
Query: 30 SILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGSTR 89
S LLELSA +D EA KREV+EKG DVNEAGFWY R+IGSKKM EKRTPLMIASLFGS R
Sbjct: 51 STLLELSAIDDIEAFKREVEEKGYDVNEAGFWYCRKIGSKKMCYEKRTPLMIASLFGSIR 230
Query: 90 VVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNKP 149
VVKY++ET V+VN A GS+ TALHCAVAGGS+ E+VK+LLDAG+D D LD +
Sbjct: 231 VVKYIIETNMVNVNMAIGSENVTALHCAVAGGSKSKFEIVKLLLDAGADVDFLDEVVRQK 410
Query: 150 MNLIAPAFNSSSKSRRKAMELLLRGGESGQLIHQEMDLELISAPFSSKEGSDKKEYPVDV 209
+++ ++SK EL+ +SG Y +D
Sbjct: 411 LSV------ANSK------ELVAEKKDSG--------------------------YAIDT 476
Query: 210 SLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPC 269
SLPDINNG++ +DEFRMYSFKVK CSR Y+HDWTECPFVHPGENARRRDPRK+PYSCVP
Sbjct: 477 SLPDINNGVFVTDEFRMYSFKVKTCSRGYTHDWTECPFVHPGENARRRDPRKFPYSCVPX 656
Query: 270 PEF 272
PEF
Sbjct: 657 PEF 665
>TC87393 similar to PIR|G84825|G84825 probable CCCH-type zinc finger protein
[imported] - Arabidopsis thaliana, partial (16%)
Length = 975
Score = 253 bits (645), Expect = 2e-67
Identities = 171/382 (44%), Positives = 204/382 (52%), Gaps = 17/382 (4%)
Frame = +1
Query: 291 ESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSV 350
ES LHP+QYRTRLCKDE C RKVCFFAH+ EELRP+YASTGSAMP
Sbjct: 1 ESLLHPSQYRTRLCKDEIRCTRKVCFFAHKHEELRPLYASTGSAMP-------------- 138
Query: 351 MSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGN-LWQNKINLNLTPPSLQLPGSRLKTA 409
S SLP+S VSTPPMSPL SPK+GN +W+NKI NLTPPSLQ LK A
Sbjct: 139 ------SQESLPISNVSTPPMSPLVADSSPKNGNYMWKNKI--NLTPPSLQ-----LKNA 279
Query: 410 LSARDFDLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLG 469
LSARD EM++L Q + ++R+ RN + P N
Sbjct: 280 LSARDLYQEMDLL------HGVSMQPSTPSQFQSMSRLQLNQNRNHVQASYPFN------ 423
Query: 470 SVDSNLLSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFD 529
NI SSP+RK S GFD
Sbjct: 424 --------------------------------------------NIVSSPMRKSSPFGFD 471
Query: 530 SSSAVAAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGK 589
SS+A+AAAVMNSRS+AFA RSQSF+DRG + ++G S ++ R++SG SDW S
Sbjct: 472 SSAAMAAAVMNSRSSAFATRSQSFMDRGVS-RQYIGASESNS---RMNSGLSDWISN--- 630
Query: 590 LDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKD-------- 641
DW DEL KL+KSASFGFRNN A SP+A +HAEPDVSWVHSLV++
Sbjct: 631 -DW----DELHKLKKSASFGFRNNMAA--ASPVARPQHAEPDVSWVHSLVQEVSSENSEI 789
Query: 642 --------DLSKEMLPPWVEQL 655
DL K+ L PW EQ+
Sbjct: 790 FGAERLHYDLYKQKLSPWTEQI 855
>AW560143 similar to PIR|G84825|G848 probable CCCH-type zinc finger protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 630
Score = 128 bits (322), Expect(2) = 2e-60
Identities = 81/133 (60%), Positives = 93/133 (69%), Gaps = 12/133 (9%)
Frame = +3
Query: 429 RQQQQQQQQQQLIEEIARISSPSFRNR----IGDLTPTNLDDLLGSVDSNLLSQLHGL-S 483
R+QQQQ QQ+ L EEIAR S PS N I + PTNL+ LL S + + L +LHGL
Sbjct: 225 RRQQQQLQQRPLSEEIARSSCPSVMNN*SYLIAEPNPTNLEHLLSSSEPH*LCELHGLYM 404
Query: 484 GPSTPTH-LQSQSRHQMMQ-----QNMNHLRSSYPSN-IPSSPVRKPSSLGFDSSSAVAA 536
P TPT + S S QM Q QNMNHLR+SYPSN +PSSPVRKPS GFDSS+AVAA
Sbjct: 405 QPMTPTQQMHSPSAMQMRQNMNMGQNMNHLRASYPSNNMPSSPVRKPSPYGFDSSAAVAA 584
Query: 537 AVMNSRSAAFAKR 549
AVMNSRSAAFAK+
Sbjct: 585 AVMNSRSAAFAKQ 623
Score = 123 bits (308), Expect(2) = 2e-60
Identities = 63/74 (85%), Positives = 68/74 (91%)
Frame = +2
Query: 352 SPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALS 411
SPL+LSSSSLPM T STPPMSPLAGS SPKSG++WQNK LNLTPPSLQLPGSRLK+ALS
Sbjct: 2 SPLSLSSSSLPMFTFSTPPMSPLAGSSSPKSGHMWQNK--LNLTPPSLQLPGSRLKSALS 175
Query: 412 ARDFDLEMEMLGLG 425
ARD DLEME+LGLG
Sbjct: 176 ARDLDLEMELLGLG 217
>TC87072 similar to PIR|D84581|D84581 probable CCCH-type zinc finger protein
[imported] - Arabidopsis thaliana, partial (47%)
Length = 1599
Score = 199 bits (506), Expect = 3e-51
Identities = 108/233 (46%), Positives = 145/233 (61%), Gaps = 5/233 (2%)
Frame = +3
Query: 200 SDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDP 259
SD + +P S+ Y +D FRM+ FK++ C+R SHDWTECPF HPGE ARRRDP
Sbjct: 213 SDSEIFPTHESVDS-----YSNDHFRMFEFKIRRCARGRSHDWTECPFSHPGEKARRRDP 377
Query: 260 RKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAH 319
RKY YS CP+FRKG+C+KGDSCE+AHGVFE WLHP++YRT+ CKD T C R VCFFAH
Sbjct: 378 RKYNYSGTSCPDFRKGSCKKGDSCEFAHGVFECWLHPSRYRTQPCKDGTSCRRPVCFFAH 557
Query: 320 RPEELRPVYASTGSAMPSPKSYSSN----AVDMSVMSPLALSSSSLPMS-TVSTPPMSPL 374
E+LR + ++PS SY + A + S + L SS +S V +PPMSP+
Sbjct: 558 TTEQLRAPTQQSPRSVPSVDSYDGSPLRLAFESSCVKTLQFMSSPGSVSPPVESPPMSPM 737
Query: 375 AGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSP 427
SL G+ N++ +L +LQL +K+ S+ ++++M GSP
Sbjct: 738 TRSLGRSVGSSSVNEMVASLR--NLQL--GTMKSLPSS--WNVQMGSPRFGSP 878
>TC86255 similar to PIR|D84581|D84581 probable CCCH-type zinc finger protein
[imported] - Arabidopsis thaliana, partial (47%)
Length = 1508
Score = 196 bits (499), Expect = 2e-50
Identities = 103/210 (49%), Positives = 128/210 (60%), Gaps = 1/210 (0%)
Frame = +3
Query: 219 YGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQ 278
Y D FRMY FK++ C+R SHDWTECP+ HPGE ARRRDPRK+ YS CP+FRKG C+
Sbjct: 333 YSCDHFRMYEFKIRRCARGRSHDWTECPYAHPGEKARRRDPRKFHYSGTACPDFRKGNCK 512
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSP 338
KGD+CE+AHGVFE WLHPA+YRT+ CKD T C R+VCFFAH PE+LR + SP
Sbjct: 513 KGDACEHAHGVFECWLHPARYRTQPCKDGTSCRRRVCFFAHTPEQLRL------NVQQSP 674
Query: 339 KSYSSNAVDMSVMSPLALSSSSL-PMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPP 397
S SV SP + S L M T++TPP SP +SP + + + NL L
Sbjct: 675 SS------SRSVNSPDSYDGSPLRQMVTITTPPESP---PMSPMASEMVASLRNLQL--- 818
Query: 398 SLQLPGSRLKTALSARDFDLEMEMLGLGSP 427
R+KT R+ + + G GSP
Sbjct: 819 ------GRMKTMPINRNVTIGSPVFGSGSP 890
>TC77687 similar to PIR|D84581|D84581 probable CCCH-type zinc finger protein
[imported] - Arabidopsis thaliana, partial (35%)
Length = 1096
Score = 173 bits (438), Expect = 2e-43
Identities = 84/161 (52%), Positives = 107/161 (66%), Gaps = 1/161 (0%)
Frame = +3
Query: 213 DINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEF 272
D+ ++ +D+FRM+ FKV+ C R SHDWT+CP+ HPGE ARRRDP+KY YS PCPEF
Sbjct: 165 DLPIHVFSTDQFRMFEFKVRKCQRGRSHDWTDCPYSHPGEKARRRDPQKYNYSGNPCPEF 344
Query: 273 RK-GACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYAST 331
RK G C KGDSC +AHGVFE WLHP++YRT+LC D T C R+VCFFAH ++LR
Sbjct: 345 RKLGNCTKGDSCHFAHGVFECWLHPSRYRTQLCNDGTLCRRRVCFFAHTIDQLRV----- 509
Query: 332 GSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPMS 372
S SP+S+ V SP ++ SS S PP++
Sbjct: 510 -SNNASPESF--------VSSPTSVLDSSPRKSRYGVPPVN 605
>TC80408 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20
{Arabidopsis thaliana}, partial (22%)
Length = 821
Score = 136 bits (343), Expect = 2e-32
Identities = 63/126 (50%), Positives = 96/126 (76%)
Frame = +3
Query: 28 NFSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGS 87
+F+ LLEL+A+ND E KR ++ + V+E G WYGRR GSK+M +E+RTPLM+A+ +GS
Sbjct: 345 SFASLLELAANNDVEGFKRLIEYDPMSVDEVGLWYGRRKGSKQMVNEQRTPLMVAATYGS 524
Query: 88 TRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGN 147
V+K + VD+N+ CG DK+TALHCA +GG+E +++ VK+LL AG+D +S+DA G+
Sbjct: 525 IDVMKLIFSLSDVDINRPCGLDKSTALHCAASGGAENAVDAVKLLLAAGADPNSVDANGD 704
Query: 148 KPMNLI 153
+P+++I
Sbjct: 705 RPIDVI 722
>BI271861
Length = 623
Score = 130 bits (328), Expect = 1e-30
Identities = 80/208 (38%), Positives = 111/208 (52%), Gaps = 23/208 (11%)
Frame = +3
Query: 478 QLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAA 537
+ HG+S S + LQS + QM HL+ +Y S +S V + F S ++A+
Sbjct: 3 KFHGISLDSAGSQLQSPTGIQMRPNMNQHLQQNYSSGHSTSSVIGSPTYRFQPSGELSAS 182
Query: 538 VMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGD 597
+N+R+AAF+KRSQSFI+RG H + SP+ P FS+W SP G LDW +G+
Sbjct: 183 ALNARAAAFSKRSQSFIERGVT-NRHSELHSPAKP-----YAFSNWGSPDGNLDWTSHGE 344
Query: 598 ELSKLRKSASFGFRNNGMAPT-GSPMAHSEHAEPDVSWVHSLVKDDLSKE---------- 646
EL+KLRKS+SF FR T + A EPDVSWV++LVKD +E
Sbjct: 345 ELNKLRKSSSFAFRTTSTPLTPAAARAQENDYEPDVSWVNTLVKDATPQESHQFSGEDQK 524
Query: 647 ------------MLPPWVEQLYIEQEQM 662
+P WVEQL ++QEQ+
Sbjct: 525 RKLQRHLNNGTDSIPAWVEQLXMDQEQI 608
>TC91525 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20
{Arabidopsis thaliana}, partial (18%)
Length = 672
Score = 106 bits (265), Expect = 3e-23
Identities = 84/213 (39%), Positives = 112/213 (52%), Gaps = 23/213 (10%)
Frame = +2
Query: 315 CFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDM-SVMSPLALSSSSLPMSTVS--TPPM 371
CFFAH PEELRP+Y STGSA+PSP+S +S+A+D + MS L S SS+ + + S TPPM
Sbjct: 2 CFFAHTPEELRPLYVSTGSAVPSPRSSTSSAMDFAAAMSMLPGSPSSMSVMSPSPFTPPM 181
Query: 372 SPLAGSLSPKSGNLWQNKINLNLTPPSLQLPG-----SRLKTALSARDFDL-EMEMLGLG 425
SP +S S Q I P+L LPG SRL+++L+ARD + + E+L
Sbjct: 182 SPSGNGISHNSVAWPQPNI------PALHLPGSNLQSSRLRSSLNARDIHMDDFELL--- 334
Query: 426 SPTRQQQQQQQQQQLIEEIARIS-------SPSFRNRIGDLTPTNLDDLLGS-------V 471
QQQQL+ E+A +S S S R+ L P+NLDDL +
Sbjct: 335 ------SDYDQQQQLLNELACLSPRHINSNSLSRSGRMKPLNPSNLDDLFSAESSSPRYA 496
Query: 472 DSNLLSQLHGLSGPSTPTHLQSQSRHQMMQQNM 504
D NL S + +PTH + QQNM
Sbjct: 497 DPNLTSTVF------SPTHKSAVFNQFQQQQNM 577
>TC76424 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20
{Arabidopsis thaliana}, partial (12%)
Length = 1429
Score = 65.1 bits (157), Expect = 8e-11
Identities = 32/66 (48%), Positives = 41/66 (61%)
Frame = +2
Query: 576 VSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWV 635
V++ +S W SP+GKLDW N DE+ KLR+S+SF NNG EPD+SWV
Sbjct: 527 VNNSWSKWESPNGKLDWAHNADEVGKLRRSSSFELGNNG-------------EEPDLSWV 667
Query: 636 HSLVKD 641
SLVK+
Sbjct: 668 QSLVKE 685
>AW585124 GP|13487732|gb ORF1 {TT virus}, partial (2%)
Length = 457
Score = 57.4 bits (137), Expect = 2e-08
Identities = 44/128 (34%), Positives = 55/128 (42%)
Frame = +3
Query: 3 SGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGFWY 62
SG KSK ++V E + G +N E +D + KRE DEK D NE G +
Sbjct: 66 SGLKSKSLLLDRVTEDKS*T--GTYNLLTPSEPLVLDDPDVPKREADEKDPDANEEGPRH 239
Query: 63 GRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGS 122
GRR G KK EKR GS R ++ G+ D N A G D + GG
Sbjct: 240 GRRTGLKKTELEKRIFSTTVLTPGSIRAAEHTALVGKADANGARGLDTVIVSYRVAVGGL 419
Query: 123 ELSLEVVK 130
E E K
Sbjct: 420 EPPSEAAK 443
>TC91252 similar to GP|9988428|dbj|BAB12694.1 putative zinc finger
transcription factor {Oryza sativa (japonica
cultivar-group)}, partial (9%)
Length = 655
Score = 40.4 bits (93), Expect = 0.002
Identities = 26/74 (35%), Positives = 37/74 (49%), Gaps = 6/74 (8%)
Frame = +1
Query: 312 RKVCFFAHRPEELRPVYASTGSAMPSP------KSYSSNAVDMSVMSPLALSSSSLPMST 365
RK+CFFAH P +LR + S P+P K S + SP + S S+
Sbjct: 28 RKICFFAHTPRQLRVLLLPPLSPSPTPPQKNNKKCCSFCHCCSNSSSPTSTLLSGPHFSS 207
Query: 366 VSTPPMSPLAGSLS 379
++PP+SPL+ S S
Sbjct: 208 SNSPPLSPLSNSNS 249
>BG647630 similar to GP|20521270|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 At5g56930, partial
(7%)
Length = 699
Score = 39.7 bits (91), Expect = 0.004
Identities = 30/118 (25%), Positives = 50/118 (41%)
Frame = +2
Query: 267 VPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRP 326
VPC + G+C KG+ C Y H + ++Y + C R C F+H+
Sbjct: 152 VPCAHYACGSCMKGNDCPYDHQL-------SKYPCSNFVSKGSCYRGRCMFSHQ------ 292
Query: 327 VYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGN 384
T +P+P +NA + SPL+ +++ STP + GS+ + N
Sbjct: 293 --VQTSQDIPTP----TNACKPELKSPLSSGNTNF-----STPLNNHGTGSVQQTNSN 433
>TC79526 weakly similar to GP|14586362|emb|CAC42893. putative protein
{Arabidopsis thaliana}, partial (72%)
Length = 797
Score = 38.1 bits (87), Expect = 0.011
Identities = 27/76 (35%), Positives = 41/76 (53%)
Frame = +3
Query: 76 RTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDA 135
RT L +A+ G +V+YL+ G D+N +K T LH A G +EVVK L+ A
Sbjct: 327 RTALHMAAANGHVNIVEYLISKG-ADLNSE-NVEKNTPLHWACLNGH---VEVVKKLIIA 491
Query: 136 GSDADSLDACGNKPMN 151
G++ L++ PM+
Sbjct: 492 GANVSVLNSYERTPMD 539
>TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 834
Score = 37.0 bits (84), Expect = 0.024
Identities = 48/164 (29%), Positives = 74/164 (44%), Gaps = 9/164 (5%)
Frame = +3
Query: 349 SVMSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNLWQNKI-NLNLTPPS----LQLPG 403
S + P + P+ S + LAG S S NL Q+ + N + PP LQ+
Sbjct: 255 SSLPPNEMQQYGEPIPGQSNNEAAKLAGG-SNASLNLPQSLVANARMLPPGNPQGLQMSQ 431
Query: 404 SRLK-TALSARDFDLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPT 462
+ L +++ R L+ + L +QQQQQQQ QQ + + +P F+ + L+
Sbjct: 432 ALLSGVSMAQRPQQLDSQQAVL----QQQQQQQQLQQNQHSLLQQQNPQFQRSL--LSAN 593
Query: 463 NLDDLLG-SVDSN--LLSQLHGLSGPSTPTHLQSQSRHQMMQQN 503
L L G +SN L + L + P LQ Q + Q +QQN
Sbjct: 594 QLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQN 725
>AW684082 homologue to PIR|T47773|T47 hypothetical protein F24I3.210 -
Arabidopsis thaliana, partial (35%)
Length = 496
Score = 36.2 bits (82), Expect = 0.042
Identities = 35/116 (30%), Positives = 51/116 (43%)
Frame = +2
Query: 38 SNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVET 97
S+D E +K V +GL+++EA L A S VVK L+E
Sbjct: 74 SSDVELVKLMVMGEGLNLDEA------------------LALPYAVENCSREVVKALLEL 199
Query: 98 GRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLI 153
G DVN G T LH A +S ++V VLLD +D + G P++++
Sbjct: 200 GAADVNFPAGPTGKTPLHIA---AEMVSPDMVAVLLDHHADPNVRTVDGVTPLDIL 358
>TC88450 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (17%)
Length = 1422
Score = 35.8 bits (81), Expect = 0.054
Identities = 44/184 (23%), Positives = 74/184 (39%), Gaps = 3/184 (1%)
Frame = +3
Query: 431 QQQQQQQQQLIEEIARIS-SPSFRNRIGDLTPTNLDDLLGSVDSNLLSQLHGLSGPSTPT 489
QQQQQ QQQ+++ +IS S S ++ T + ++ S Q S ++ T
Sbjct: 117 QQQQQTQQQVVDNNQQISGSVSTMSQFVSATQPQSPPPMQALSSLCHQQSFSDSNVNSST 296
Query: 490 HLQSQSRHQMMQQNMNHLRSSYPSNIP--SSPVRKPSSLGFDSSSAVAAAVMNSRSAAFA 547
+ S H +M + H SS ++P SS V +S G+ S +++S ++
Sbjct: 297 TIVS-PLHSIMGSSFPHDESSLLMSLPRTSSWVPVQNSTGWPSKRIAVDPLLSSGASQCI 473
Query: 548 KRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDELSKLRKSAS 607
+ + ++ P P S N P L +GVN D S L +
Sbjct: 474 LPQVEQLGQARNSMSQNAITLPPFPGRECSIDQEGSNDPQSNLLFGVNIDPSSLLLHNGM 653
Query: 608 FGFR 611
F+
Sbjct: 654 SNFK 665
>TC84988 similar to GP|14018368|emb|CAC38358. zinc finger protein {Pisum
sativum}, partial (28%)
Length = 656
Score = 34.7 bits (78), Expect = 0.12
Identities = 12/23 (52%), Positives = 16/23 (69%)
Frame = +1
Query: 269 CPEFRKGACQKGDSCEYAHGVFE 291
C +FR G C+ G++C YAHG E
Sbjct: 505 CTKFRFGTCRNGENCNYAHGADE 573
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.129 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,929,945
Number of Sequences: 36976
Number of extensions: 325886
Number of successful extensions: 2213
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 1799
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2003
length of query: 664
length of database: 9,014,727
effective HSP length: 102
effective length of query: 562
effective length of database: 5,243,175
effective search space: 2946664350
effective search space used: 2946664350
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0204.2


