

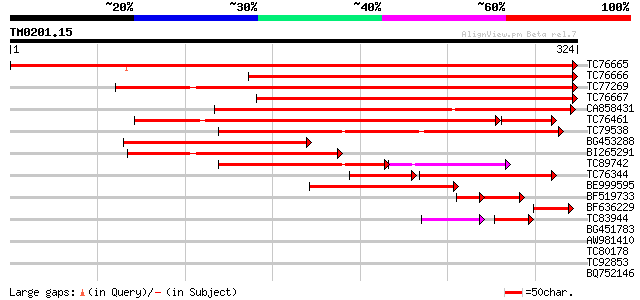
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.15
(324 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76665 homologue to SP|P17067|CAHC_PEA Carbonic anhydrase chlo... 543 e-155
TC76666 homologue to GP|8569256|pdb|1EKJ|G Chain G The X-Ray Cr... 361 e-100
TC77269 similar to SP|P27141|CAHC_TOBAC Carbonic anhydrase chlo... 343 4e-95
TC76667 homologue to GP|8569256|pdb|1EKJ|G Chain G The X-Ray Cr... 327 3e-90
CA858431 similar to PIR|T09797|T09 carbonate dehydratase (EC 4.2... 306 5e-84
TC76461 homologue to PIR|T09570|T09570 carbonate dehydratase (EC... 234 2e-69
TC79538 similar to GP|21594039|gb|AAM65957.1 carbonate dehydrata... 209 2e-54
BG453288 homologue to SP|P17067|CAHC Carbonic anhydrase chlorop... 179 1e-45
BI265291 similar to GP|606805|gb|AA carbonic anhydrase {Flaveria... 152 2e-37
TC89742 weakly similar to GP|21594039|gb|AAM65957.1 carbonate de... 101 1e-31
TC76344 similar to PIR|T09570|T09570 carbonate dehydratase (EC 4... 93 6e-29
BE999595 GP|13359451|db putative senescence-associated protein {... 112 1e-25
BF519733 homologue to GP|8954289|gb|A carbonic anhydrase {Vigna ... 50 2e-06
BF636229 homologue to GP|16226255|gb| AT3g01500/F4P13_5 {Arabido... 45 5e-05
TC83944 homologue to PIR|T09570|T09570 carbonate dehydratase (EC... 38 1e-04
BG451783 similar to GP|14334470|gb| putative carbonic anhydrase ... 33 0.19
AW981410 similar to GP|9743350|gb| F21J9.27 {Arabidopsis thalian... 33 0.19
TC80178 similar to GP|4512677|gb|AAD21731.1| unknown protein {Ar... 31 0.74
TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Ar... 31 0.74
BQ752146 similar to GP|7958612|gb|A PalH {Emericella nidulans}, ... 31 0.74
>TC76665 homologue to SP|P17067|CAHC_PEA Carbonic anhydrase chloroplast
precursor (EC 4.2.1.1) (Carbonate dehydratase). [Garden
pea], complete
Length = 1352
Score = 543 bits (1398), Expect = e-155
Identities = 279/331 (84%), Positives = 308/331 (92%), Gaps = 7/331 (2%)
Frame = +1
Query: 1 MSTSTINGFYLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSS-FPSLIQDRPVFAAPA 59
MSTS+INGF LSS+SP KTS+K+VTLRP+V AS +S SSSSS+S FPSLIQD+PVFA+ +
Sbjct: 55 MSTSSINGFSLSSLSPTKTSIKKVTLRPIVSASLNSSSSSSSTSNFPSLIQDKPVFASSS 234
Query: 60 -PII-PTL--DMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSS-DGIPSS 114
PII P L +MGK Y+EAIEELQKLLREK ELKATAAEKVEQITAQLGTT+S DG+P+S
Sbjct: 235 SPIITPVLREEMGKGYDEAIEELQKLLREKTELKATAAEKVEQITAQLGTTASADGVPTS 414
Query: 115 E-ASDRIKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEA 173
+ AS+RIKTGFL+FKKEKYD PALYGELAKGQ+PP+MVFACSDSRVCPSHVLDFQPGEA
Sbjct: 415 DQASERIKTGFLHFKKEKYDTKPALYGELAKGQAPPFMVFACSDSRVCPSHVLDFQPGEA 594
Query: 174 FVVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTT 233
FVVRNVANMVPPYDQAK++G+G+AIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDG
Sbjct: 595 FVVRNVANMVPPYDQAKYAGTGSAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGAY 774
Query: 234 STDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNK 293
STDFIE+WV IGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLL+YPFVREGLVNK
Sbjct: 775 STDFIEEWVKIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLTYPFVREGLVNK 954
Query: 294 TLALKGGYYDFVKGSFELWSLQFDLASSFSV 324
TLALKGGYYDFVKGSFELW L+F L+S+FSV
Sbjct: 955 TLALKGGYYDFVKGSFELWGLEFGLSSTFSV 1047
>TC76666 homologue to GP|8569256|pdb|1EKJ|G Chain G The X-Ray
Crystallographic Structure Of Beta Carbonic Anhydrase
From The C3 Dicot Pisum, partial (85%)
Length = 858
Score = 361 bits (927), Expect = e-100
Identities = 172/188 (91%), Positives = 183/188 (96%)
Frame = +3
Query: 137 ALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKFSGSGA 196
ALYGELAKGQ+PP+MVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAK++G+G+
Sbjct: 9 ALYGELAKGQAPPFMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKYAGTGS 188
Query: 197 AIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKH 256
AIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDG STDFIE+WV IGLPAKAKVKAKH
Sbjct: 189 AIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGAYSTDFIEEWVKIGLPAKAKVKAKH 368
Query: 257 GDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQF 316
GDAPFGELCTHCEKEAVNVSLGNLL+YPFVREGLVNKTLALKGGYYDFVKGSFELW L+F
Sbjct: 369 GDAPFGELCTHCEKEAVNVSLGNLLTYPFVREGLVNKTLALKGGYYDFVKGSFELWGLEF 548
Query: 317 DLASSFSV 324
L+S+FSV
Sbjct: 549 GLSSTFSV 572
>TC77269 similar to SP|P27141|CAHC_TOBAC Carbonic anhydrase chloroplast
precursor (EC 4.2.1.1) (Carbonate dehydratase). [Common
tobacco], partial (64%)
Length = 1230
Score = 343 bits (881), Expect = 4e-95
Identities = 169/265 (63%), Positives = 211/265 (78%), Gaps = 1/265 (0%)
Frame = +2
Query: 61 IIPTLDM-GKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDR 119
I+ DM G+ +E++I L +LL+EK EL AA K++++TA+L ++G +R
Sbjct: 176 IVSKEDMAGETFEDSIATLTRLLKEKAELGDIAAVKIKELTAEL---EANGSKPFNPDER 346
Query: 120 IKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNV 179
I++GF+ FK EK+ KNP LYGELAKGQSP +MVFACSDSRVCPSH+LDFQPGEAFVVRN+
Sbjct: 347 IRSGFVSFKTEKFLKNPELYGELAKGQSPKFMVFACSDSRVCPSHILDFQPGEAFVVRNI 526
Query: 180 ANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIE 239
ANMVPP+D+ K+SG+GAAIEYAVLHLKV NIVVIGHS CGGIKGL+S P DGTT++DFIE
Sbjct: 527 ANMVPPFDKTKYSGAGAAIEYAVLHLKVENIVVIGHSCCGGIKGLMSIPDDGTTASDFIE 706
Query: 240 DWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKG 299
WV I PA++KVK + F E CT+CEKEAVNVSLGNLL+YPFVR+G+V K+LALKG
Sbjct: 707 QWVQICNPARSKVKLETSSLSFAEQCTNCEKEAVNVSLGNLLTYPFVRDGVVKKSLALKG 886
Query: 300 GYYDFVKGSFELWSLQFDLASSFSV 324
+Y+FV G+FELW L F+L S S+
Sbjct: 887 AHYNFVNGTFELWDLNFNLLPSVSI 961
>TC76667 homologue to GP|8569256|pdb|1EKJ|G Chain G The X-Ray
Crystallographic Structure Of Beta Carbonic Anhydrase
From The C3 Dicot Pisum, partial (83%)
Length = 1017
Score = 327 bits (839), Expect = 3e-90
Identities = 158/183 (86%), Positives = 168/183 (91%)
Frame = +3
Query: 142 LAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKFSGSGAAIEYA 201
L K + P SDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAK++G+G+AIEYA
Sbjct: 12 LPKAKPPRLWCLHASDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKYAGTGSAIEYA 191
Query: 202 VLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPF 261
VLHLKVSNIVVIGHSACGGIKGLLSFPFDG STDFIE+WV IGLPAKAKVKAKHGDAPF
Sbjct: 192 VLHLKVSNIVVIGHSACGGIKGLLSFPFDGAYSTDFIEEWVKIGLPAKAKVKAKHGDAPF 371
Query: 262 GELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQFDLASS 321
GELCTHCEKEAVNVSLGNLL+YPFVREGLVNKTLALKGGYYDFVKGSFELW L+F L+S+
Sbjct: 372 GELCTHCEKEAVNVSLGNLLTYPFVREGLVNKTLALKGGYYDFVKGSFELWGLEFGLSST 551
Query: 322 FSV 324
FSV
Sbjct: 552 FSV 560
Score = 33.5 bits (75), Expect = 0.11
Identities = 13/15 (86%), Positives = 15/15 (99%)
Frame = +2
Query: 141 ELAKGQSPPYMVFAC 155
ELAKGQ+PP+MVFAC
Sbjct: 8 ELAKGQAPPFMVFAC 52
>CA858431 similar to PIR|T09797|T09 carbonate dehydratase (EC 4.2.1.1) 1b -
Populus tremula x Populus tremuloides, partial (63%)
Length = 903
Score = 306 bits (785), Expect = 5e-84
Identities = 146/206 (70%), Positives = 177/206 (85%)
Frame = +1
Query: 118 DRIKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVR 177
+RI++GF YFKKEK+ K P LYGELAKGQSP ++VFACSDSRVCPS+VL+FQPGEAFVVR
Sbjct: 22 ERIRSGFEYFKKEKFLKKPELYGELAKGQSPKFLVFACSDSRVCPSYVLNFQPGEAFVVR 201
Query: 178 NVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDF 237
N+ANMVPPYD++K+SG+GAAIEYAV+HLKV NIVVIGHS CGGIKGL+S P DGTT++DF
Sbjct: 202 NIANMVPPYDKSKYSGTGAAIEYAVVHLKVENIVVIGHSCCGGIKGLMSIPDDGTTASDF 381
Query: 238 IEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLAL 297
IE WV I PAK+KVK + ++ F E CT+ EKEAVNVSLGNLL+YPFVR+ +V KT+AL
Sbjct: 382 IEQWVQICNPAKSKVK-EASNSDFSEQCTNLEKEAVNVSLGNLLTYPFVRDAVVKKTIAL 558
Query: 298 KGGYYDFVKGSFELWSLQFDLASSFS 323
KG +YDFV G+FELW + F ++ S S
Sbjct: 559 KGAHYDFVNGAFELWDIDFKISPSVS 636
>TC76461 homologue to PIR|T09570|T09570 carbonate dehydratase (EC 4.2.1.1) -
alfalfa, complete
Length = 1504
Score = 234 bits (597), Expect(2) = 2e-69
Identities = 119/209 (56%), Positives = 146/209 (68%)
Frame = +1
Query: 72 EEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYFKKEK 131
E AIE+L+KLLREK EL A K+EQ+ +L + P A RI GF YFK
Sbjct: 610 ELAIEQLKKLLREKEELNGVATAKIEQLIVELQGCHPN--PIEPADQRIIDGFTYFKLNN 783
Query: 132 YDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKF 191
++KNP LY LAKGQSP +MVFACSDSRV PS +L+FQPGEAF+VRN+ANMVPP++Q ++
Sbjct: 784 FNKNPELYDRLAKGQSPKFMVFACSDSRVSPSVILNFQPGEAFMVRNIANMVPPFNQLRY 963
Query: 192 SGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAK 251
SG GA +EYA+ LKV NI+VIGHS CGGI L++ P DG+ DFI+DWV IGL +K K
Sbjct: 964 SGVGATLEYAITALKVENILVIGHSRCGGISRLMNHPEDGSAPYDFIDDWVKIGLSSKVK 1143
Query: 252 VKAKHGDAPFGELCTHCEKEAVNVSLGNL 280
V +H F E C CE E+VN SL NL
Sbjct: 1144VLKEHERCDFKEQCKFCEMESVNNSLVNL 1230
Score = 46.2 bits (108), Expect(2) = 2e-69
Identities = 19/31 (61%), Positives = 23/31 (73%)
Frame = +3
Query: 282 SYPFVREGLVNKTLALKGGYYDFVKGSFELW 312
+YP+V + NK LAL GGYYDFV G F+LW
Sbjct: 1233 AYPYVDREIRNKNLALLGGYYDFVSGEFKLW 1325
>TC79538 similar to GP|21594039|gb|AAM65957.1 carbonate dehydratase-like
protein {Arabidopsis thaliana}, partial (57%)
Length = 1260
Score = 209 bits (531), Expect = 2e-54
Identities = 103/197 (52%), Positives = 137/197 (69%)
Frame = +2
Query: 120 IKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNV 179
+K FL FKK Y +NP + LAK Q+P +MV AC+DSRVCPS++L FQPG+AF +RNV
Sbjct: 407 LKDRFLSFKKNVYMENPEQFESLAKVQTPKFMVIACADSRVCPSNILGFQPGDAFTIRNV 586
Query: 180 ANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIE 239
AN+VP ++ S + AA+E+AV L V NI+V+GHS CGGI+ L+ DG+TS FI+
Sbjct: 587 ANLVPTFESGP-SETNAALEFAVNTLLVENILVVGHSCCGGIRALMGMQDDGSTS--FIK 757
Query: 240 DWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKG 299
WV G AK K K + F CTHCEKE++N SL NLLSYP+++E + N+ L++ G
Sbjct: 758 SWVIHGKNAKVKTKVSASNLDFDHQCTHCEKESINHSLVNLLSYPWIKEKVENEELSIHG 937
Query: 300 GYYDFVKGSFELWSLQF 316
GYYDFV SFE W+L +
Sbjct: 938 GYYDFVNCSFEKWTLDY 988
>BG453288 homologue to SP|P17067|CAHC Carbonic anhydrase chloroplast
precursor (EC 4.2.1.1) (Carbonate dehydratase). [Garden
pea], partial (33%)
Length = 524
Score = 179 bits (454), Expect = 1e-45
Identities = 90/109 (82%), Positives = 101/109 (92%), Gaps = 2/109 (1%)
Frame = +3
Query: 66 DMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSS-DGIPSSE-ASDRIKTG 123
+MGK Y+EAIEELQKLLREK ELKATAAEKVEQITAQLGTT+S DG+P+S+ AS+RIKTG
Sbjct: 198 EMGKGYDEAIEELQKLLREKTELKATAAEKVEQITAQLGTTASADGVPTSDQASERIKTG 377
Query: 124 FLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGE 172
FL+FKKEKYD PALYGELAKGQ+PP+MVFACS SRVCPSHVL+F PGE
Sbjct: 378 FLHFKKEKYDTKPALYGELAKGQAPPFMVFACSXSRVCPSHVLNFXPGE 524
>BI265291 similar to GP|606805|gb|AA carbonic anhydrase {Flaveria bidentis},
partial (24%)
Length = 412
Score = 152 bits (383), Expect = 2e-37
Identities = 73/123 (59%), Positives = 96/123 (77%)
Frame = +3
Query: 68 GKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYF 127
G+ +E++I L +LL+EK EL AA K++++TA+L ++G +RI++GF+ F
Sbjct: 51 GETFEDSIATLTRLLKEKAELGDIAAVKIKELTAEL---EANGSKPFNPDERIRSGFVSF 221
Query: 128 KKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYD 187
K EK+ KNP LYGELAKGQSP +MVFACSDS VCPSH+LDFQPGEAFVVRN+ANMVPP+D
Sbjct: 222 KTEKFLKNPELYGELAKGQSPKFMVFACSDSXVCPSHILDFQPGEAFVVRNIANMVPPFD 401
Query: 188 QAK 190
+ K
Sbjct: 402 KTK 410
>TC89742 weakly similar to GP|21594039|gb|AAM65957.1 carbonate
dehydratase-like protein {Arabidopsis thaliana}, partial
(31%)
Length = 846
Score = 101 bits (252), Expect(2) = 1e-31
Identities = 51/98 (52%), Positives = 68/98 (69%)
Frame = +2
Query: 120 IKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNV 179
+K FL FK +KY K Y LA+ Q P +MV AC+DSRVCPS++L FQPGE F++RN+
Sbjct: 350 MKQRFLNFKNQKYMKELDHYESLAEAQYPKFMVIACADSRVCPSNILGFQPGEVFMIRNI 529
Query: 180 ANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSA 217
AN+VP S AA+++AV L+V NI+VIGHS+
Sbjct: 530 ANLVPMMKNGP-SECNAALQFAVTTLQVENILVIGHSS 640
Score = 52.4 bits (124), Expect(2) = 1e-31
Identities = 27/70 (38%), Positives = 41/70 (58%)
Frame = +3
Query: 217 ACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVS 276
AC GI+ L+ D T ++I +WV+ G + + K+ H + C CEKE++N S
Sbjct: 639 ACAGIEALMKMQED-TEPRNYIHNWVANGKSCQIENKSCHISSLL*STCRFCEKESINQS 815
Query: 277 LGNLLSYPFV 286
L NLLSYP++
Sbjct: 816 LLNLLSYPWI 845
>TC76344 similar to PIR|T09570|T09570 carbonate dehydratase (EC 4.2.1.1) -
alfalfa, partial (51%)
Length = 777
Score = 93.2 bits (230), Expect(2) = 6e-29
Identities = 44/78 (56%), Positives = 53/78 (67%)
Frame = +2
Query: 235 TDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKT 294
+DFI+DWV IGL +K KV +H F E C CE E+VN SL NL +YP+V + NK
Sbjct: 209 SDFIDDWVKIGLSSKVKVLKEHERCDFKEQCKFCEMESVNNSLVNLKTYPYVDREIRNKN 388
Query: 295 LALKGGYYDFVKGSFELW 312
LAL GGYYDFV G F+LW
Sbjct: 389 LALLGGYYDFVSGEFKLW 442
Score = 52.0 bits (123), Expect(2) = 6e-29
Identities = 23/38 (60%), Positives = 29/38 (75%)
Frame = +3
Query: 195 GAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGT 232
GA +EYA+ LKV NI+VIGHS CGGI L++ P DG+
Sbjct: 3 GATLEYAITALKVENILVIGHSRCGGISRLMNHPEDGS 116
>BE999595 GP|13359451|db putative senescence-associated protein {Pisum
sativum}, partial (37%)
Length = 579
Score = 112 bits (281), Expect = 1e-25
Identities = 51/85 (60%), Positives = 66/85 (77%)
Frame = +3
Query: 172 EAFVVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDG 231
EAF+VRN+ANMVPP++Q ++SG GA +EYA+ LKV NI+VIGHS CGGI L++ P DG
Sbjct: 324 EAFMVRNIANMVPPFNQLRYSGVGATLEYAITALKVENILVIGHSRCGGISRLMNHPEDG 503
Query: 232 TTSTDFIEDWVSIGLPAKAKVKAKH 256
+ DFI+DWV IGL +K KV +H
Sbjct: 504 SAPYDFIDDWVKIGLSSKVKVLKEH 578
>BF519733 homologue to GP|8954289|gb|A carbonic anhydrase {Vigna radiata},
partial (11%)
Length = 582
Score = 49.7 bits (117), Expect = 2e-06
Identities = 23/25 (92%), Positives = 25/25 (100%)
Frame = +3
Query: 270 KEAVNVSLGNLLSYPFVREGLVNKT 294
+EAVNVSLGNLL+YPFVREGLVNKT
Sbjct: 507 QEAVNVSLGNLLTYPFVREGLVNKT 581
Score = 42.4 bits (98), Expect = 2e-04
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +3
Query: 256 HGDAPFGELCTHCEKE 271
HGDAPFGELCTHCEKE
Sbjct: 3 HGDAPFGELCTHCEKE 50
>BF636229 homologue to GP|16226255|gb| AT3g01500/F4P13_5 {Arabidopsis
thaliana}, partial (8%)
Length = 285
Score = 44.7 bits (104), Expect = 5e-05
Identities = 18/23 (78%), Positives = 21/23 (91%)
Frame = +3
Query: 300 GYYDFVKGSFELWSLQFDLASSF 322
GYYDFVKGSFELW L+F L+S+F
Sbjct: 3 GYYDFVKGSFELWGLEFGLSSTF 71
>TC83944 homologue to PIR|T09570|T09570 carbonate dehydratase (EC 4.2.1.1) -
alfalfa, partial (19%)
Length = 763
Score = 37.7 bits (86), Expect(2) = 1e-04
Identities = 18/36 (50%), Positives = 21/36 (58%)
Frame = +1
Query: 236 DFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKE 271
DFI+DWV IGL +K KV +H F E CE E
Sbjct: 550 DFIDDWVKIGLSSKVKVLKEHECNDFKEQFKFCEME 657
Score = 24.6 bits (52), Expect(2) = 1e-04
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +3
Query: 278 GNLLSYPFVREGLVNKTLALKG 299
GNL +YP+V + NK LA G
Sbjct: 654 GNLKTYPYVDR*IRNKNLATTG 719
>BG451783 similar to GP|14334470|gb| putative carbonic anhydrase {Arabidopsis
thaliana}, partial (6%)
Length = 645
Score = 32.7 bits (73), Expect = 0.19
Identities = 12/17 (70%), Positives = 16/17 (93%)
Frame = +3
Query: 150 YMVFACSDSRVCPSHVL 166
+MV AC+DSRVCPS++L
Sbjct: 594 FMVIACADSRVCPSNIL 644
>AW981410 similar to GP|9743350|gb| F21J9.27 {Arabidopsis thaliana},
partial (22%)
Length = 595
Score = 32.7 bits (73), Expect = 0.19
Identities = 16/23 (69%), Positives = 20/23 (86%)
Frame = +3
Query: 20 SLKRVTLRPLVVASASSPSSSSS 42
S+K+VTLRP+V AS +S SSSSS
Sbjct: 3 SIKKVTLRPIVSASLNSSSSSSS 71
>TC80178 similar to GP|4512677|gb|AAD21731.1| unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 946
Score = 30.8 bits (68), Expect = 0.74
Identities = 20/48 (41%), Positives = 26/48 (53%), Gaps = 6/48 (12%)
Frame = -1
Query: 7 NGFYLSSISPAKTS------LKRVTLRPLVVASASSPSSSSSSSFPSL 48
NGF+LS S + L T + + S+SSPSS+SS S PSL
Sbjct: 721 NGFFLSL*SKTSQASDK*GLLSDKTFSSIFLCSSSSPSSASSFSLPSL 578
>TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 761
Score = 30.8 bits (68), Expect = 0.74
Identities = 34/119 (28%), Positives = 50/119 (41%), Gaps = 4/119 (3%)
Frame = -2
Query: 11 LSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAPIIPTLDMGKN 70
LSS S +SL + + +S+SS SSSSS S PS + P++ T +G
Sbjct: 484 LSSSSALNSSLSLPSSSLVSSSSSSSSSSSSSFSLPSSSLSLSLPLVSLPLVSTGILGFG 305
Query: 71 YEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTS----SDGIPSSEASDRIKTGFL 125
E A + + LLR + + + A TS SDG + D I + L
Sbjct: 304 GEGATKLIVALLRVLDTILLISIPSSSLARAFFLFTSLISFSDGFEDKPSLDSISSNSL 128
>BQ752146 similar to GP|7958612|gb|A PalH {Emericella nidulans}, partial (5%)
Length = 641
Score = 30.8 bits (68), Expect = 0.74
Identities = 17/33 (51%), Positives = 20/33 (60%)
Frame = +3
Query: 28 PLVVASASSPSSSSSSSFPSLIQDRPVFAAPAP 60
P+ +S+SS SSSSS S S D P AAP P
Sbjct: 465 PMSRSSSSSSSSSSSRSGSSTETDEPSLAAPVP 563
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,881,624
Number of Sequences: 36976
Number of extensions: 147413
Number of successful extensions: 1680
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1228
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1485
length of query: 324
length of database: 9,014,727
effective HSP length: 96
effective length of query: 228
effective length of database: 5,465,031
effective search space: 1246027068
effective search space used: 1246027068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0201.15


