

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.13
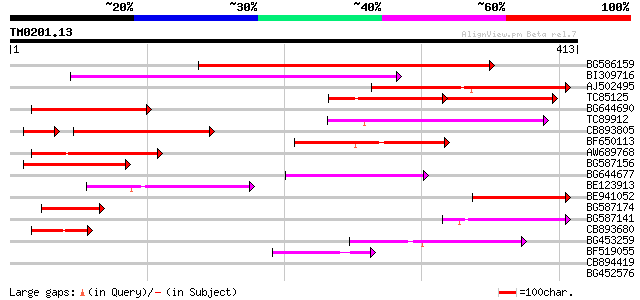
(413 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 212 2e-55
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 162 2e-40
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 137 8e-33
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 78 8e-31
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 126 2e-29
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 115 3e-26
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 97 3e-23
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 96 2e-20
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 87 2e-17
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 78 5e-15
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 73 2e-13
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 72 4e-13
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 64 8e-11
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 60 1e-09
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 52 5e-07
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 49 5e-06
BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, ... 47 1e-05
BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thalian... 47 2e-05
CB894419 weakly similar to GP|10177935|db copia-type polyprotein... 39 0.003
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 33 0.030
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 212 bits (539), Expect = 2e-55
Identities = 103/217 (47%), Positives = 146/217 (66%), Gaps = 1/217 (0%)
Frame = +1
Query: 138 IQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGM 197
++V Q EG YI Q KY +LL++F M +S +++ P+ P C L K++ KV Y+ +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 198 IGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYK 257
+G L+YL A+RPD+++ + L +RF + P E H+ AVKR+LRYL GT NLG+MYK+ K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 258 LSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQM 317
L Y D+DYAGD +RKSTSG L S VSW+SK+Q + LST +AE+I+AA C+ Q
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 318 LWMKH*LEDYQILES-NIPIYCDNTAAISLSKNPILH 353
+WM+ LE +S +I +YCDN + I LSKNP+LH
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 162 bits (410), Expect = 2e-40
Identities = 91/241 (37%), Positives = 134/241 (54%)
Frame = +2
Query: 45 VFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDII 104
V +L+KS+YGLKQA R WY +LS L+ +++ D +LF K + +YVDDI+
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 105 FGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNM 164
+ S + + F++ +G L+YFLG++V ++ +G ++Q KYT ELL+
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGN 379
Query: 165 LESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSD 224
L TP + L D + YR +IG L+YLT +RPDI F+V ++F S
Sbjct: 380 LAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSK 559
Query: 225 PRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLG 284
P++ H A R+L+YLK GL Y TS KLS + D+D+A T RKS +G FLG
Sbjct: 560 PQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFLG 739
Query: 285 S 285
S
Sbjct: 740 S 742
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 137 bits (345), Expect = 8e-33
Identities = 68/147 (46%), Positives = 101/147 (68%), Gaps = 2/147 (1%)
Frame = +2
Query: 264 ADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH* 323
+D+AGD RKSTSG LG+ +SW+SK+Q +A STAEAEYI++ C+TQ +W++
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 324 LEDYQILESNIP--IYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVD 381
LE E N P IYCDN +AI+LSKNP+ H R+KHI++++H IR+ + + +++++
Sbjct: 182 LEVMHH-EQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCP 358
Query: 382 TDHQWADIFTKPLAEDRFNFILKNLNM 408
T+ + ADIFTKPL + F + K L M
Sbjct: 359 TEEKIADIFTKPLKIESFYKLKKMLGM 439
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 77.8 bits (190), Expect(2) = 8e-31
Identities = 40/87 (45%), Positives = 58/87 (65%)
Frame = +1
Query: 233 VKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWAS 292
VKRI+RY+KGT+ + + + SE + GY D+D+AGD +RKST+G L VSW S
Sbjct: 1 VKRIMRYIKGTSGVAVCFGG-SELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 293 KRQSTIALSTAEAEYISAAICSTQMLW 319
K Q+ +ALST EAEY++A + + L+
Sbjct: 178 KLQTVVALSTTEAEYMAAYLKHARKLF 258
Score = 73.9 bits (180), Expect(2) = 8e-31
Identities = 29/84 (34%), Positives = 55/84 (64%)
Frame = +3
Query: 316 QMLWMKH*LEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVL 375
+ +WM+ +E+ + I +YCD+ +A+ +++NP HSR KHI ++YHF+R+ V++G +
Sbjct: 249 EAIWMQRLMEELGHKQEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVEEGSV 428
Query: 376 LLKFVDTDHQWADIFTKPLAEDRF 399
++ + T+ AD TK + D+F
Sbjct: 429 DMQKIHTNDNLADAMTKSINTDKF 500
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 126 bits (316), Expect = 2e-29
Identities = 58/87 (66%), Positives = 72/87 (82%)
Frame = -2
Query: 17 SAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFV 76
SAF+NG + EEV+V QPPGFED + P+HVF+L K+LYGLKQAPRAWYERLS FLL+N F
Sbjct: 262 SAFINGDLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFK 83
Query: 77 RGKVDTTLFCKTYKDDILIVQIYVDDI 103
RGK+D TLF + ++LI+Q+YVDDI
Sbjct: 82 RGKIDNTLFLLKRE*ELLIIQVYVDDI 2
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 115 bits (288), Expect = 3e-26
Identities = 58/163 (35%), Positives = 97/163 (58%), Gaps = 2/163 (1%)
Frame = +1
Query: 232 AVKRILRYLKGTTNLGLMYKKTSEYK--LSGYCDADYAGDRTERKSTSGNCQFLGSNLVS 289
A+K +L+YL + L Y K ++ + L GY DADYAG+ RKS SG L +S
Sbjct: 4 ALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTIS 183
Query: 290 WASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQILESNIPIYCDNTAAISLSKN 349
W + +QS + LST +AEYI+ +W+K + + I + + I+CD+ +AI L+ +
Sbjct: 184 WKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEYVKIHCDSQSAIHLANH 363
Query: 350 PILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTK 392
+ H R KHI+++ HFIRD ++ ++++ + ++ AD+FTK
Sbjct: 364 QVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 96.7 bits (239), Expect(2) = 3e-23
Identities = 45/104 (43%), Positives = 72/104 (68%), Gaps = 1/104 (0%)
Frame = +3
Query: 47 KLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDD-ILIVQIYVDDIIF 105
++K++LYGLKQAPRAWY R+ ++ + F + + TLF K + ILI+ +YVDD+IF
Sbjct: 462 RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIF 641
Query: 106 GSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYI 149
++++ +EF + M+ EF MS +G++ YFLG++V Q +G YI
Sbjct: 642 IGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVEVTQNEKGIYI 773
Score = 29.6 bits (65), Expect(2) = 3e-23
Identities = 13/26 (50%), Positives = 20/26 (76%)
Frame = +2
Query: 11 WRLRM*SAFLNGYISEEVYVHQPPGF 36
++L + SAFL G ++EEV+V QP G+
Sbjct: 362 YQLDVKSAFLYGELNEEVFVDQPQGY 439
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 96.3 bits (238), Expect = 2e-20
Identities = 50/116 (43%), Positives = 71/116 (61%), Gaps = 3/116 (2%)
Frame = +1
Query: 208 RPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMY---KKTSEYKLSGYCDA 264
RPDI +SV + ++F DPR+ HL A RILRY++GT GL++ K+ Y+L Y D+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 265 DYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWM 320
D+ GD R+STSG +SW +K+Q ALS+ EAEYI+ + Q LW+
Sbjct: 301 DWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWL 459
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 86.7 bits (213), Expect = 2e-17
Identities = 45/95 (47%), Positives = 60/95 (62%)
Frame = +1
Query: 17 SAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFV 76
+AFLNG++ EEVY+ QP GFE K V KL KSLYGLKQAPRAWYE L+S ++ F
Sbjct: 394 NAFLNGFLQEEVYMSQPQGFEAANK-SLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFT 570
Query: 77 RGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQS 111
+ + D +L + + IYVDDI+ ++ S
Sbjct: 571 KSRCDPSLLIYNQNGACIYLXIYVDDILITGSSAS 675
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 78.2 bits (191), Expect = 5e-15
Identities = 36/78 (46%), Positives = 54/78 (69%)
Frame = -1
Query: 11 WRLRM*SAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFL 70
W++ + +AFL G + +EVY++ PPG E K +V +LKK++YGLKQ+PRAWY +LS+ L
Sbjct: 243 WQMDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTL 64
Query: 71 LENEFVRGKVDTTLFCKT 88
F + ++D TLF T
Sbjct: 63 NGRGFRKSELDHTLFTLT 10
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 72.8 bits (177), Expect = 2e-13
Identities = 39/104 (37%), Positives = 58/104 (55%)
Frame = -3
Query: 202 LYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGY 261
+ LT P+I FS++L +R+ S P H +K I +YLKG ++GL Y K L GY
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 262 CDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEA 305
+A Y D + +S +G G+ ++SW S + STIA S+ A
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 72.0 bits (175), Expect = 4e-13
Identities = 42/125 (33%), Positives = 67/125 (53%), Gaps = 3/125 (2%)
Frame = +1
Query: 57 QAPRAWYERLSSFLLENEFVRGKVDTTLFCK---TYKDDILIVQIYVDDIIFGSANQSLC 113
Q+PR W++R + + + +++ + D +F K T K ILIV YVDDI +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIV--YVDDIFLTGDHGK*I 174
Query: 114 KEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTP 173
K ++ EFE+ +G LKYFLG++V + +G+ I Q KY +LLK+ M+ + P
Sbjct: 175 KRLKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Query: 174 MHPTC 178
C
Sbjct: 355 YGCNC 369
Score = 36.2 bits (82), Expect = 0.024
Identities = 20/57 (35%), Positives = 32/57 (56%)
Frame = +2
Query: 165 LESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARF 221
L+ ++TPM T L D V + Y+ ++G L+YL+ +RPDI F V ++F
Sbjct: 329 LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQF 499
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 64.3 bits (155), Expect = 8e-11
Identities = 30/71 (42%), Positives = 43/71 (60%)
Frame = +2
Query: 338 CDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAED 397
CD +A L+ NP+ HSR KHI + HF+RD VQ+G L ++ V T Q AD TKPL++
Sbjct: 29 CDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLADCLTKPLSKS 208
Query: 398 RFNFILKNLNM 408
R + + +
Sbjct: 209 RHQLLRNKIGV 241
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 60.5 bits (145), Expect = 1e-09
Identities = 26/46 (56%), Positives = 34/46 (73%)
Frame = -1
Query: 24 ISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSF 69
+ E++Y+ QP GF K DHV KL+KSLYGLKQ+PR WY+R S+
Sbjct: 246 LEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSY 109
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 731
Score = 51.6 bits (122), Expect = 5e-07
Identities = 31/95 (32%), Positives = 51/95 (53%), Gaps = 2/95 (2%)
Frame = +3
Query: 316 QMLWMKH*LED--YQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKG 373
Q +W++ L + ++ E + I DN + I+L++NP+ H R HI +YHFIR+ V+ G
Sbjct: 135 QAMWLQDLLSEVTWEPCEE-VVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRECVENG 311
Query: 374 VLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 408
+ ++ V + A I TK L F I + M
Sbjct: 312 QVEVEHVPGEKHRAYI*TKALGRIIFREIRYYIGM 416
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 48.5 bits (114), Expect = 5e-06
Identities = 25/44 (56%), Positives = 31/44 (69%)
Frame = -2
Query: 17 SAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPR 60
+AFL G + E++Y+HQP GF E V KLKKS+YGLKQ PR
Sbjct: 464 TAFLRGDLVEDIYMHQPEGFS*E-VGKMVGKLKKSMYGLKQGPR 336
>BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 657
Score = 47.0 bits (110), Expect = 1e-05
Identities = 36/132 (27%), Positives = 65/132 (48%), Gaps = 3/132 (2%)
Frame = -2
Query: 248 LMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIA--LSTAEA 305
+++K+ + + Y + AG +R STSG FLG N+V +Q+ +A +
Sbjct: 530 IVFKRE*KLSMEVYXNTVCAGWIVDRGSTSGY*MFLGGNMVE*---KQNVVAR*VQRHNF 360
Query: 306 EYISAAICSTQMLWMKH*LEDYQI-LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYH 364
E S + +K L+D I + + ++ +N ++ NP+ H R KHIE+ H
Sbjct: 359 ELCSQGL*RVMDEELKIKLDDLIINYKDPMTLF*NNNFVSRIAHNPVQHYRTKHIEIDQH 180
Query: 365 FIRDYVQKGVLL 376
FI + + G++L
Sbjct: 179 FIIEKLYSGLIL 144
>BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thaliana}, partial
(30%)
Length = 675
Score = 46.6 bits (109), Expect = 2e-05
Identities = 22/75 (29%), Positives = 44/75 (58%)
Frame = -3
Query: 192 KLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYK 251
KL ++G+LLY+T + PD+ FS++ ++F P + + +K+++R+ K L K
Sbjct: 640 KLVHSIVGALLYITVTCPDLSFSINKPSQFMHKPTQINFQQLKKVMRHPK------LTIK 479
Query: 252 KTSEYKLSGYCDADY 266
K + ++ + DAD+
Sbjct: 478 KLFDLQIYAFSDADW 434
>CB894419 weakly similar to GP|10177935|db copia-type polyprotein
{Arabidopsis thaliana}, partial (2%)
Length = 170
Score = 39.3 bits (90), Expect = 0.003
Identities = 21/55 (38%), Positives = 29/55 (52%)
Frame = +3
Query: 153 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTAS 207
K ++LKKF M S TP+ L +E +V Y+ +IGSL YLTA+
Sbjct: 6 KCASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT 170
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like
protein {Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 32.7 bits (73), Expect(2) = 0.030
Identities = 26/103 (25%), Positives = 49/103 (47%)
Frame = +2
Query: 63 YERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQA 122
Y +L S L+ + + D ++F ++ I +Y+DDI+F + S + +
Sbjct: 245 Y*KLLSTLISLGYKQSPNDHSIF--SFGRRFTIFLVYLDDIVFARDDHSETQLVKSHLDK 418
Query: 123 EFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNML 165
F + +G L Y +G+++ + Q KYT ELL++ L
Sbjct: 419 NFIIIGLGTLHY-VGVKIA*SES*IIDDQCKYTIELLEESGQL 544
Score = 21.9 bits (45), Expect(2) = 0.030
Identities = 10/21 (47%), Positives = 12/21 (56%)
Frame = +3
Query: 45 VFKLKKSLYGLKQAPRAWYER 65
V K S+YGLK A R Y +
Sbjct: 165 VCKFHNSIYGLKHAHRQ*YSK 227
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,110,945
Number of Sequences: 36976
Number of extensions: 178463
Number of successful extensions: 1026
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1009
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1016
length of query: 413
length of database: 9,014,727
effective HSP length: 99
effective length of query: 314
effective length of database: 5,354,103
effective search space: 1681188342
effective search space used: 1681188342
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0201.13


