

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
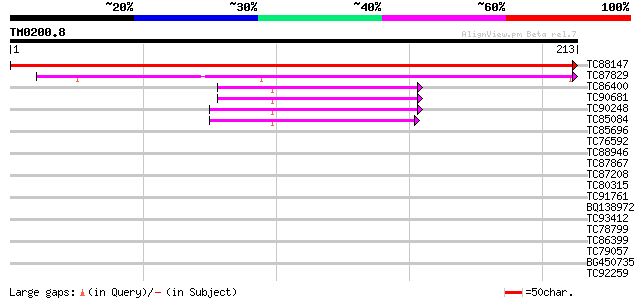
Query= TM0200.8
(213 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88147 similar to SP|O64628|14P_ARATH Protein At2g18990. [Mouse... 377 e-105
TC87829 similar to GP|21592510|gb|AAM64460.1 ATP-binding protein... 143 6e-35
TC86400 weakly similar to GP|21595244|gb|AAM66084.1 thioredoxin ... 53 1e-07
TC90681 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin ... 46 9e-06
TC90248 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin ... 45 2e-05
TC85084 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin ... 45 2e-05
TC85696 similar to SP|Q43636|THIH_RICCO Thioredoxin H-type (TRX-... 39 0.001
TC76592 similar to GP|1388088|gb|AAC49358.1| thioredoxin m {Pisu... 39 0.002
TC88946 similar to GP|21592332|gb|AAM64283.1 thioredoxin putati... 38 0.003
TC87867 similar to GP|15594012|emb|CAC69854. putative thioredoxi... 38 0.003
TC87208 similar to SP|Q43636|THIH_RICCO Thioredoxin H-type (TRX-... 37 0.007
TC80315 similar to PIR|B96796|B96796 thioredoxin-like protein 4... 36 0.010
TC91761 similar to GP|21554313|gb|AAM63418.1 thioredoxin-like pr... 35 0.016
BQ138972 similar to SP|P29429|THIO_ Thioredoxin. [Aspergillus ni... 35 0.028
TC93412 similar to GP|15594012|emb|CAC69854. putative thioredoxi... 33 0.081
TC78799 weakly similar to SP|P96132|THIO_THIRO Thioredoxin (TRX)... 33 0.11
TC86399 similar to PIR|T00710|T00710 thioredoxin homolog F22O13.... 31 0.40
TC79057 similar to GP|14581677|gb|AAK64512.1 Hsp70 interacting p... 31 0.40
BG450735 similar to PIR|T00710|T00 thioredoxin homolog F22O13.5 ... 30 0.69
TC92259 weakly similar to GP|10177335|dbj|BAB10684. nuclear matr... 30 0.69
>TC88147 similar to SP|O64628|14P_ARATH Protein At2g18990. [Mouse-ear cress]
{Arabidopsis thaliana}, partial (86%)
Length = 1081
Score = 377 bits (967), Expect = e-105
Identities = 188/213 (88%), Positives = 202/213 (94%)
Frame = +1
Query: 1 MDKGKIQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNR 60
MD+GKIQEVIE QVLTV QAVEDKID+EI+AL+RLD DDLE+LRERRL QMKKMAEKR+R
Sbjct: 70 MDRGKIQEVIENQVLTVAQAVEDKIDEEIAALERLDDDDLEALRERRLQQMKKMAEKRSR 249
Query: 61 WISLGHGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQHIETRFVK 120
WISLGHGDY+E+PSEKDFFSVVKAS+RVVCHFFRENWPCKV DKHL +LAKQHIETRFVK
Sbjct: 250 WISLGHGDYTEIPSEKDFFSVVKASERVVCHFFRENWPCKVVDKHLSILAKQHIETRFVK 429
Query: 121 INAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVI 180
INAEKSPFLAEKLKI VLPT+ALIKN KVDDYVVGFDELGGTDDFSTE LEERLAKAQVI
Sbjct: 430 INAEKSPFLAEKLKIIVLPTIALIKNTKVDDYVVGFDELGGTDDFSTEVLEERLAKAQVI 609
Query: 181 FLEGESSIHQARSTAQSKRSVRQATRADSSDSE 213
FLEGESSIH+ARS AQ+KRSVRQ+T ADSSDSE
Sbjct: 610 FLEGESSIHRARSGAQTKRSVRQSTTADSSDSE 708
>TC87829 similar to GP|21592510|gb|AAM64460.1 ATP-binding protein-like
protein {Arabidopsis thaliana}, partial (98%)
Length = 1127
Score = 143 bits (360), Expect = 6e-35
Identities = 79/206 (38%), Positives = 121/206 (58%), Gaps = 3/206 (1%)
Frame = +3
Query: 11 EKQVLTVVQAVEDK-IDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHGDY 69
+K++L+ +A + I++E+ D +D +LE L R+ KK AEKR W GHG++
Sbjct: 234 QKELLSEEKAAQSSSINEEVDLDDLMDDPELEKLHADRIAAFKKEAEKREAWKKKGHGEF 413
Query: 70 SELPSEKDFFSVVKASDRVVCHFF-RENWPCKVADKHLGVLAKQHIETRFVKINAEKSPF 128
E+ +E DF V S++V+CHF+ +E + CK+ DKHL L+ +HI+T+F++++AE +PF
Sbjct: 414 REV-TEGDFLGEVTGSEKVICHFYHKEFYRCKIMDKHLKSLSTKHIDTKFIRLDAENAPF 590
Query: 129 LAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGESSI 188
KL I LP + L + D +VGF +LG DDFST LE L K +I + +
Sbjct: 591 FVAKLAIKTLPCVILFRQGVAVDRLVGFQDLGAKDDFSTRALEVLLIKKGIIAEKKDVDE 770
Query: 189 HQARSTAQSKRSVRQATRADS-SDSE 213
+RSVR + ADS SDS+
Sbjct: 771 EDQEYDESKRRSVRSSAAADSDSDSD 848
>TC86400 weakly similar to GP|21595244|gb|AAM66084.1 thioredoxin
{Arabidopsis thaliana}, partial (82%)
Length = 746
Score = 52.8 bits (125), Expect = 1e-07
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 2/79 (2%)
Frame = +2
Query: 79 FSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKIT 136
F+ +K S R+V F W PCK+ + L +A + + F+KI+ ++ +A++ K+
Sbjct: 212 FNELKDSPRLVVIDFSATWCGPCKMMEPILQAMANEFTDVEFIKIDVDELSDVAQEFKVQ 391
Query: 137 VLPTLALIKNAKVDDYVVG 155
+PT L+KN K D VVG
Sbjct: 392 AMPTFLLLKNGKEVDKVVG 448
>TC90681 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin H {Populus
tremula x Populus tremuloides}, partial (60%)
Length = 637
Score = 46.2 bits (108), Expect = 9e-06
Identities = 24/79 (30%), Positives = 43/79 (54%), Gaps = 2/79 (2%)
Frame = +2
Query: 79 FSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKIT 136
F K +++++ F W PCKV D + A ++ + F+K++ ++ +++ ++
Sbjct: 167 FEAFKETNKLMVIDFTAKWCGPCKVMDPVMKDFAAKYTDVEFIKLDVDELMGVSQTFQVH 346
Query: 137 VLPTLALIKNAKVDDYVVG 155
LPT LIK KV D VVG
Sbjct: 347 SLPTFLLIKKGKVVDKVVG 403
>TC90248 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin H {Populus
tremula x Populus tremuloides}, partial (63%)
Length = 660
Score = 45.4 bits (106), Expect = 2e-05
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Frame = +1
Query: 76 KDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKL 133
K F K +++++ F W PCK D + LA ++ + F+KI+ ++ +A
Sbjct: 163 KAHFEASKVTNKLMVIDFTATWCGPCKYMDPIIKELAAKYKDVEFIKIDVDELMDVASAF 342
Query: 134 KITVLPTLALIKNAKVDDYVVG 155
++ +PT L+K KV + VVG
Sbjct: 343 QVQAMPTFILLKKGKVVEKVVG 408
>TC85084 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin H {Populus
tremula x Populus tremuloides}, partial (41%)
Length = 601
Score = 45.4 bits (106), Expect = 2e-05
Identities = 23/81 (28%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Frame = +3
Query: 76 KDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKL 133
K +F K +++++ F W PCK D + A ++I+ F+KI+ ++ ++++
Sbjct: 138 KAYFDASKETNKLMVIEFTAAWCGPCKYMDPIIQDFAAKYIKVDFIKIDVDELMSISQEF 317
Query: 134 KITVLPTLALIKNAKVDDYVV 154
++ +PT L+K KV D VV
Sbjct: 318 QVQAMPTFILMKKGKVVDKVV 380
>TC85696 similar to SP|Q43636|THIH_RICCO Thioredoxin H-type (TRX-H). [Castor
bean] {Ricinus communis}, partial (94%)
Length = 714
Score = 39.3 bits (90), Expect = 0.001
Identities = 26/110 (23%), Positives = 51/110 (45%), Gaps = 2/110 (1%)
Frame = +2
Query: 48 LHQMKKMAEKRNRWISLGHGDYSELPSEKDFFSVVKASDRVVCHFFRENW--PCKVADKH 105
+ + KKMA + I + + + K+ S +++ F +W PC+
Sbjct: 83 VRRKKKMAAEEGHVIGV-----HTVEAWKEHLEKGNGSKKLIVVDFTASWCGPCRFIAPI 247
Query: 106 LGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVG 155
L +AK+ F+K++ ++ +AE+ + +PT +K K+ D VVG
Sbjct: 248 LAEIAKKLPNVTFLKVDVDELKTVAEEWAVDAMPTFLFLKEGKLVDKVVG 397
>TC76592 similar to GP|1388088|gb|AAC49358.1| thioredoxin m {Pisum sativum},
partial (87%)
Length = 912
Score = 38.5 bits (88), Expect = 0.002
Identities = 29/107 (27%), Positives = 55/107 (51%), Gaps = 5/107 (4%)
Frame = +1
Query: 54 MAEKRNRWISLGHGDYSELPSEKD--FFSVVKASDRVVCHFFRENW--PCKVADKHLGVL 109
+A +++R+I +E+ + D + +V ASD V F W PC++ + L
Sbjct: 265 VAYRKSRFICNAREAVNEVGAVTDSSWNELVLASDTPVLVDFWAPWCGPCRMIAPIIDEL 444
Query: 110 AKQHI-ETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVG 155
AK++ + K+N +++P +A K I +PT+ KN + + V+G
Sbjct: 445 AKEYAGKISCYKLNTDENPNIATKYGIRSIPTVLFFKNGEKKESVIG 585
>TC88946 similar to GP|21592332|gb|AAM64283.1 thioredoxin putative
{Arabidopsis thaliana}, partial (61%)
Length = 761
Score = 38.1 bits (87), Expect = 0.003
Identities = 25/78 (32%), Positives = 41/78 (52%), Gaps = 3/78 (3%)
Frame = +3
Query: 74 SEKDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIET-RFVKINAEKSPFLA 130
+E F V S+R V F W PC++ + +A+++ + VKI+ + +P L
Sbjct: 213 NESQFNVTVLKSERPVLVEFVATWCGPCRLITPAMESIAQEYADRLTVVKIDHDANPQLI 392
Query: 131 EKLKITVLPTLALIKNAK 148
E+ K+ LPTL L KN +
Sbjct: 393 EEYKVYGLPTLILFKNGE 446
>TC87867 similar to GP|15594012|emb|CAC69854. putative thioredoxin m2 {Pisum
sativum}, partial (79%)
Length = 816
Score = 38.1 bits (87), Expect = 0.003
Identities = 24/79 (30%), Positives = 42/79 (52%), Gaps = 3/79 (3%)
Frame = +3
Query: 80 SVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHI-ETRFVKINAEKSPFLAEKLKIT 136
S+V S+ V F W PC++ + LAK++ + + K+N ++SP +A + I
Sbjct: 291 SLVIESETPVLVEFWAPWCGPCRMIHPIIDELAKEYAGKLKCYKLNTDESPSVATRYGIR 470
Query: 137 VLPTLALIKNAKVDDYVVG 155
+PT+ KN + D V+G
Sbjct: 471 SIPTVIFFKNGEKKDTVIG 527
>TC87208 similar to SP|Q43636|THIH_RICCO Thioredoxin H-type (TRX-H). [Castor
bean] {Ricinus communis}, partial (94%)
Length = 682
Score = 36.6 bits (83), Expect = 0.007
Identities = 20/73 (27%), Positives = 38/73 (51%), Gaps = 2/73 (2%)
Frame = +3
Query: 85 SDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLA 142
S +++ F +W PC+ L +AK+ E F+K++ ++ +A++ + +PT
Sbjct: 123 SKKLIVVDFTASWCGPCRFIAPILAEIAKKIPEVIFLKVDIDEVKSVAKEWSVEAMPTFL 302
Query: 143 LIKNAKVDDYVVG 155
+K K D VVG
Sbjct: 303 FLKEGKEVDKVVG 341
>TC80315 similar to PIR|B96796|B96796 thioredoxin-like protein 49720-48645
[imported] - Arabidopsis thaliana, partial (70%)
Length = 802
Score = 36.2 bits (82), Expect = 0.010
Identities = 27/81 (33%), Positives = 38/81 (46%), Gaps = 3/81 (3%)
Frame = +2
Query: 78 FFSVVKASDRVVCHFFRENW--PCKVADKHLG-VLAKQHIETRFVKINAEKSPFLAEKLK 134
F ++ SD+ V F W PC+ L V A+ + + VKI+ EK P +A K
Sbjct: 188 FDDLLANSDKPVFVDFYATWCGPCQFMVPVLEEVSARLQDQIQIVKIDTEKYPSIANKYN 367
Query: 135 ITVLPTLALIKNAKVDDYVVG 155
I LPT + K+ K D G
Sbjct: 368 IEALPTFIIFKDGKPFDRFEG 430
>TC91761 similar to GP|21554313|gb|AAM63418.1 thioredoxin-like protein
{Arabidopsis thaliana}, partial (55%)
Length = 613
Score = 35.4 bits (80), Expect = 0.016
Identities = 22/92 (23%), Positives = 45/92 (48%), Gaps = 3/92 (3%)
Frame = +2
Query: 60 RWISLGHGDYSELPSEKDFFSVV-KASDRVVCHFFRENW--PCKVADKHLGVLAKQHIET 116
+W + ++ S ++F + + +A DR+V F W C+ L A++H E
Sbjct: 245 KWWEKNASNMIDIHSTQEFLNALSQAEDRLVIVEFYGTWCASCRALFPKLCRTAEEHPEI 424
Query: 117 RFVKINAEKSPFLAEKLKITVLPTLALIKNAK 148
F+K+N +++ + + L + VLP + A+
Sbjct: 425 IFLKVNFDENKPMCKSLNVKVLPYFHFYRGAE 520
>BQ138972 similar to SP|P29429|THIO_ Thioredoxin. [Aspergillus nidulans]
{Emericella nidulans}, partial (73%)
Length = 653
Score = 34.7 bits (78), Expect = 0.028
Identities = 18/58 (31%), Positives = 29/58 (49%)
Frame = +1
Query: 98 PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVG 155
PCKV + + + RF K++ ++ P +A++L I +PT L K VVG
Sbjct: 253 PCKVIAPQVVKFSSTYPNARFYKLDVDEVPDVAQELGIRAMPTFLLFKGGDKVAEVVG 426
>TC93412 similar to GP|15594012|emb|CAC69854. putative thioredoxin m2 {Pisum
sativum}, partial (47%)
Length = 624
Score = 33.1 bits (74), Expect = 0.081
Identities = 17/59 (28%), Positives = 33/59 (55%), Gaps = 1/59 (1%)
Frame = +1
Query: 98 PCKVADKHLGVLAKQHI-ETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVG 155
PC++ + LA Q+ + + K+N ++SP A + I +PT+ + K+ + D V+G
Sbjct: 34 PCRMITPIIDELAIQYTGKLKCYKLNTDESPSTATRYGIRSIPTVMIFKDGEKKDTVIG 210
>TC78799 weakly similar to SP|P96132|THIO_THIRO Thioredoxin (TRX)
(Fragment). {Thiocapsa roseopersicina}, partial (37%)
Length = 734
Score = 32.7 bits (73), Expect = 0.11
Identities = 23/87 (26%), Positives = 41/87 (46%), Gaps = 4/87 (4%)
Frame = +2
Query: 74 SEKDFFSVVKASDRVVCHFFRENW---PCKVADKHLGVLAKQHI-ETRFVKINAEKSPFL 129
+++ F S+V S +V F W C + LA + + F K+N +++P++
Sbjct: 212 TDETFGSIVPMSKNLVLVEFYAPWCGQECINIHSIMVELANDYAGKVDFYKLNIDENPYI 391
Query: 130 AEKLKITVLPTLALIKNAKVDDYVVGF 156
+ I LPT+ IK D +VG+
Sbjct: 392 TNRYGIQNLPTVVFIKYGMQRDRLVGY 472
>TC86399 similar to PIR|T00710|T00710 thioredoxin homolog F22O13.5 -
Arabidopsis thaliana, partial (61%)
Length = 1568
Score = 30.8 bits (68), Expect = 0.40
Identities = 24/96 (25%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Frame = +2
Query: 57 KRNRWISLGHG-DYSELPSEKDFF-SVVKASDR-VVCHFFRENWP-CKVADKHLGVLAKQ 112
K +W GH + E+ S +D S++ A D+ V+ FF CK + A+
Sbjct: 443 KVQKWWEKGHQPNMREVTSAQDLVDSLLNAGDKLVIVDFFSPGCGGCKALHPKICQFAEM 622
Query: 113 HIETRFVKINAEKSPFLAEKLKITVLPTLALIKNAK 148
+ + F+++N E+ + L + VLP + A+
Sbjct: 623 NPDVEFLQVNYEEHKSMCYSLNVHVLPFFRFYRGAQ 730
>TC79057 similar to GP|14581677|gb|AAK64512.1 Hsp70 interacting
protein/thioredoxin chimera {Vitis labrusca}, partial
(84%)
Length = 1335
Score = 30.8 bits (68), Expect = 0.40
Identities = 13/41 (31%), Positives = 26/41 (62%)
Frame = +3
Query: 118 FVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDE 158
F+K++ +++ +A + ++ +PT +KN K D VVG D+
Sbjct: 978 FLKVDIDEAVDVAARWNVSSVPTFFFVKNGKEVDSVVGADK 1100
>BG450735 similar to PIR|T00710|T00 thioredoxin homolog F22O13.5 -
Arabidopsis thaliana, partial (38%)
Length = 635
Score = 30.0 bits (66), Expect = 0.69
Identities = 22/71 (30%), Positives = 33/71 (45%), Gaps = 2/71 (2%)
Frame = +1
Query: 80 SVVKASDR-VVCHFFRENWP-CKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITV 137
S+V A D VV F+ CK + +A+ H F+K+N E+ + E L I V
Sbjct: 310 SLVNAGDTLVVVDFYSPGCGGCKALHPKICQIAELHPSVIFLKVNYEELRSMCESLHIHV 489
Query: 138 LPTLALIKNAK 148
LP + A+
Sbjct: 490 LPFFRFYRGAE 522
>TC92259 weakly similar to GP|10177335|dbj|BAB10684. nuclear matrix
constituent protein 1 (NMCP1)-like {Arabidopsis
thaliana}, partial (7%)
Length = 630
Score = 30.0 bits (66), Expect = 0.69
Identities = 18/50 (36%), Positives = 25/50 (50%)
Frame = +1
Query: 12 KQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRW 61
K V T V+ DK+ +E L + + DL L R + MKKMA + W
Sbjct: 172 KAVSTFVKNERDKLREEKENLRKQYTHDLGLLASERENFMKKMAHEHAEW 321
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,893,573
Number of Sequences: 36976
Number of extensions: 57024
Number of successful extensions: 316
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 315
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 315
length of query: 213
length of database: 9,014,727
effective HSP length: 92
effective length of query: 121
effective length of database: 5,612,935
effective search space: 679165135
effective search space used: 679165135
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0200.8


