

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
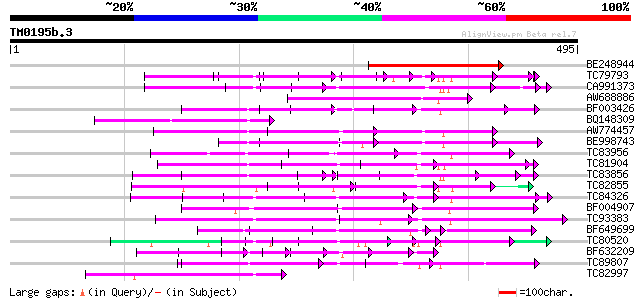
Query= TM0195b.3
(495 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE248944 similar to GP|11994509|dbj gb|AAF27119.1~gene_id:F4B12.... 212 3e-55
TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Ar... 114 1e-25
CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imp... 109 2e-24
AW688886 similar to GP|3258568|gb|A Unknown protein {Arabidopsis... 97 1e-20
BF003426 similar to PIR|F96760|F96 hypothetical protein T9L24.39... 97 1e-20
BQ148309 weakly similar to GP|6721172|gb| hypothetical protein {... 93 2e-19
AW774457 weakly similar to GP|10176973|dbj gb|AAF19552.1~gene_id... 92 5e-19
BE998743 similar to GP|22128591|gb| fertility restorer-like prot... 84 1e-16
TC83956 weakly similar to GP|15982931|gb|AAL09812.1 AT3g53700/F4... 84 2e-16
TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Ar... 82 6e-16
TC83856 weakly similar to PIR|B96656|B96656 unknown protein 419... 80 1e-15
TC82855 weakly similar to PIR|T45829|T45829 hypothetical protein... 69 2e-15
TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein... 79 4e-15
BF004907 weakly similar to GP|8953393|emb putative protein {Arab... 78 7e-15
TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16... 73 2e-13
BF649699 similar to GP|9757911|dbj gb|AAF19552.1~gene_id:MMN10.1... 73 3e-13
TC80520 weakly similar to PIR|T02562|T02562 probable salt-induci... 72 4e-13
BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01... 71 8e-13
TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [i... 69 3e-12
TC82997 homologue to GP|7293564|gb|AAF48937.1| CG7884 gene produ... 69 3e-12
>BE248944 similar to GP|11994509|dbj gb|AAF27119.1~gene_id:F4B12.11~similar
to unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 357
Score = 212 bits (539), Expect = 3e-55
Identities = 100/118 (84%), Positives = 105/118 (88%)
Frame = +2
Query: 314 VFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNS 373
VF DMKER C PNVATYNSLIKHLCKIRRMEKVYELVEDMER+ G C+PN VTYS LL S
Sbjct: 2 VFHDMKERDCLPNVATYNSLIKHLCKIRRMEKVYELVEDMERRSGDCLPNGVTYSYLLQS 181
Query: 374 LKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDR 431
LK PEEVP VLERMERNGC++SDDI NLVLRLYMKWD+ DGLRKTWDEMERNG GPDR
Sbjct: 182 LKAPEEVPAVLERMERNGCAMSDDICNLVLRLYMKWDDLDGLRKTWDEMERNGLGPDR 355
Score = 40.8 bits (94), Expect = 0.001
Identities = 28/114 (24%), Positives = 54/114 (46%)
Frame = +2
Query: 242 VWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDA 301
V+ D+ C P++ TY + IK L K ++ +L M +C P+ V + ++ +
Sbjct: 2 VFHDMKERDCLPNVATYNSLIKHLCKIRRMEKVYELVEDMERRSGDCLPNGVTYSYLLQS 181
Query: 302 LCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMER 355
L + VP LE M+ GC + N +++ K ++ + + ++MER
Sbjct: 182 LKAPEEVPAVLE---RMERNGCAMSDDICNLVLRLYMKWDDLDGLRKTWDEMER 334
>TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Arabidopsis
thaliana}, partial (7%)
Length = 1580
Score = 114 bits (284), Expect = 1e-25
Identities = 70/244 (28%), Positives = 125/244 (50%), Gaps = 3/244 (1%)
Frame = +1
Query: 222 TWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGM 281
T+N +++G+C++ ++AK ++ + PD+ +Y+ I K K A+ LF+ M
Sbjct: 562 TYNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEAMNLFKEM 741
Query: 282 WNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIR 341
+ N PDVV + +ID L R+ AL++ M +RG PN+ TYNS++ LCK
Sbjct: 742 HRK--NIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDALCKTH 915
Query: 342 RMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGP---EEVPGVLERMERNGCSLSDDI 398
+++K L+ + K P+ TYS L+ L E+ V E + G +L+ D
Sbjct: 916 QVDKAIALLTKFKDK--GFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGHNLNVDT 1089
Query: 399 YNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMT 458
Y ++++ + + +ME NGC PD ++Y I+I ++ + A + REM
Sbjct: 1090YTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIIILSLFKKDENDMAEKLLREMI 1269
Query: 459 SKGM 462
++G+
Sbjct: 1270ARGL 1281
Score = 113 bits (282), Expect = 2e-25
Identities = 70/251 (27%), Positives = 135/251 (52%), Gaps = 3/251 (1%)
Frame = +1
Query: 179 GLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHE 238
G+ + + +L+ C K V A+++F++ A+ ++ DI+++++++NG+C + E
Sbjct: 541 GIKPNFVTYNSLMDGYCLVKEVNKAKSIFNTMAQG-GVNPDIQSYSIMINGFCKIKKFDE 717
Query: 239 AKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCI 298
A ++K++ PD+ TY++ I L+K G++ AL+L M + G P++ N I
Sbjct: 718 AMNLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRG--VPPNICTYNSI 891
Query: 299 IDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKG 358
+DALC +V +A+ + K++G +P+++TY+ LIK LC+ ++E ++ E + K
Sbjct: 892 LDALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGH 1071
Query: 359 SCMPNAVTYSCLLNS--LKGP-EEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGL 415
+ N TY+ ++ ++G E +L +ME NGC Y +++ K D D
Sbjct: 1072NL--NVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIIILSLFKKDENDMA 1245
Query: 416 RKTWDEMERNG 426
K EM G
Sbjct: 1246EKLLREMIARG 1278
Score = 72.0 bits (175), Expect = 5e-13
Identities = 42/135 (31%), Positives = 70/135 (51%)
Frame = +2
Query: 219 DIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLF 278
D T+ + G C+ G +A ++A D +Y T I L K G+ AL L
Sbjct: 26 DTITFTTLSKGLCLKGQIQQAFLFHDKVVALGFHFDQISYGTLIHGLCKVGETRAALDLL 205
Query: 279 RGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLC 338
+ + +G +P+VV+ N IID++C K V EA ++F +M +G P+V TY++LI C
Sbjct: 206 QRV--DGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLFSEMVSKGISPDVVTYSALISGFC 379
Query: 339 KIRRMEKVYELVEDM 353
+ +++ +L M
Sbjct: 380 ILGKLKDAIDLFNKM 424
Score = 70.1 bits (170), Expect = 2e-12
Identities = 46/168 (27%), Positives = 82/168 (48%)
Frame = +1
Query: 118 NEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQ 177
+ ++D L K R Q+ D+M R N T++++L H+V++AI++ +
Sbjct: 778 SSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDALCKTHQVDKAIALLTKFKD 957
Query: 178 FGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAH 237
G D+ + L+ LC+ +EDA +F + + ++ T+ +++ G+CV G +
Sbjct: 958 KGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGH-NLNVDTYTIMIQGFCVEGLFN 1134
Query: 238 EAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEG 285
EA + + + C PD TY I +L KK + A KL R M G
Sbjct: 1135EALALLSKMEDNGCIPDAKTYEIIILSLFKKDENDMAEKLLREMIARG 1278
Score = 62.4 bits (150), Expect = 4e-10
Identities = 39/148 (26%), Positives = 71/148 (47%)
Frame = +2
Query: 183 DLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRV 242
D F TL LC ++ A LFH K H D ++ +++G C +G A +
Sbjct: 26 DTITFTTLSKGLCLKGQIQQA-FLFHDKVVALGFHFDQISYGTLIHGLCKVGETRAALDL 202
Query: 243 WKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDAL 302
+ + + +P++ Y T I ++ K + A LF M ++G + PDVV + +I
Sbjct: 203 LQRVDGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLFSEMVSKGIS--PDVVTYSALISGF 376
Query: 303 CFKKRVPEALEVFQDMKERGCEPNVATY 330
C ++ +A+++F M +P+V T+
Sbjct: 377 CILGKLKDAIDLFNKMILENIKPDVYTF 460
Score = 57.4 bits (137), Expect = 1e-08
Identities = 45/179 (25%), Positives = 74/179 (41%), Gaps = 38/179 (21%)
Frame = +1
Query: 321 RGCEPNVATYNSL-----------------------------------IKHLCKIRRMEK 345
+G +PN TYNSL I CKI++ ++
Sbjct: 538 KGIKPNFVTYNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDE 717
Query: 346 VYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVL---ERMERNGCSLSDDIYNLV 402
L ++M RK + +P+ VTYS L++ L + L ++M G + YN +
Sbjct: 718 AMNLFKEMHRK--NIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSI 891
Query: 403 LRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKG 461
L K D + + G PD +Y+I+I G ++GK++DA + F + KG
Sbjct: 892 LDALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKG 1068
Score = 55.1 bits (131), Expect = 6e-08
Identities = 34/120 (28%), Positives = 58/120 (48%)
Frame = +2
Query: 253 PDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEAL 312
PD T+ T K L KG++ A + G + D + +I LC AL
Sbjct: 23 PDTITFTTLSKGLCLKGQIQQAFLFHDKVVALGFHF--DQISYGTLIHGLCKVGETRAAL 196
Query: 313 EVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLN 372
++ Q + +PNV YN++I +CK++ + + ++L +M K P+ VTYS L++
Sbjct: 197 DLLQRVDGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLFSEMVSK--GISPDVVTYSALIS 370
Score = 43.9 bits (102), Expect = 1e-04
Identities = 39/169 (23%), Positives = 68/169 (40%), Gaps = 1/169 (0%)
Frame = +2
Query: 290 PDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYEL 349
PD + + LC K ++ +A + G + +Y +LI LCK+ +L
Sbjct: 23 PDTITFTTLSKGLCLKGQIQQAFLFHDKVVALGFHFDQISYGTLIHGLCKVGETRAALDL 202
Query: 350 VEDMERKKGSCM-PNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMK 408
++R G+ + PN V Y+ +++S+ C + + N L+
Sbjct: 203 ---LQRVDGNLVQPNVVMYNTIIDSM-----------------CKVK--LVNEAFDLF-- 310
Query: 409 WDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREM 457
EM G PD +Y+ +I G GK+KDA+ F +M
Sbjct: 311 -----------SEMVSKGISPDVVTYSALISGFCILGKLKDAIDLFNKM 424
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/75 (29%), Positives = 39/75 (51%)
Frame = +1
Query: 399 YNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMT 458
YN ++ Y + + ++ M + G PD +SY+IMI+G + K +AM F+EM
Sbjct: 565 YNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEAMNLFKEMH 744
Query: 459 SKGMVAEPRTEKLVI 473
K ++ + T +I
Sbjct: 745 RKNIIPDVVTYSSLI 789
>CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 756
Score = 109 bits (273), Expect = 2e-24
Identities = 69/239 (28%), Positives = 127/239 (52%), Gaps = 3/239 (1%)
Frame = +2
Query: 189 TLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMA 248
+L+ C K V A+++ ++ ++ ++ DI+++N++++G+C + EA ++K++
Sbjct: 41 SLMDGYCLVKEVNKAKSILYTMSQR-GVNPDIQSYNILIDGFCKIKKVDEAMNLFKEMHH 217
Query: 249 SKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRV 308
PD+ TY + I L K GK+ ALKL M + G PD++ + I+DALC +V
Sbjct: 218 KHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRG--VPPDIITYSSILDALCKNHQV 391
Query: 309 PEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYS 368
+A+ + +K++G PN+ TY LI LCK R+E + + ED+ K + N TY+
Sbjct: 392 DKAIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYNITVN--TYT 565
Query: 369 CLLNSL--KGP-EEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMER 424
+++ KG +E +L +M+ N C Y +++R D D K + + R
Sbjct: 566 VMIHGFCNKGLFDEALTLLSKMKDNSCIPDAVTYEIIIRSLFDKDENDKAEKLREMITR 742
Score = 101 bits (252), Expect = 6e-22
Identities = 65/247 (26%), Positives = 127/247 (51%), Gaps = 3/247 (1%)
Frame = +2
Query: 220 IKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFR 279
+ T+ +++G+C++ ++AK + + PD+ +Y I K K+ A+ LF+
Sbjct: 26 VVTYCSLMDGYCLVKEVNKAKSILYTMSQRGVNPDIQSYNILIDGFCKIKKVDEAMNLFK 205
Query: 280 GMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCK 339
M ++ + PDVV N +ID LC ++ AL++ +M +RG P++ TY+S++ LCK
Sbjct: 206 EMHHK--HIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSILDALCK 379
Query: 340 IRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL-KGP--EEVPGVLERMERNGCSLSD 396
+++K L+ + K PN TY+ L++ L KG E+ + E + G +++
Sbjct: 380 NHQVDKAIALLTKL--KDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYNITV 553
Query: 397 DIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFRE 456
+ Y +++ + D +M+ N C PD +Y I+I ++ + D RE
Sbjct: 554 NTYTVMIHGFCNKGLFDEALTLLSKMKDNSCIPDAVTYEIIIRSLFDKDE-NDKAEKLRE 730
Query: 457 MTSKGMV 463
M ++G++
Sbjct: 731 MITRGLL 751
Score = 88.6 bits (218), Expect = 5e-18
Identities = 57/230 (24%), Positives = 109/230 (46%), Gaps = 3/230 (1%)
Frame = +2
Query: 247 MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKK 306
M +P + TY + + ++ A + M G N PD+ N +ID C K
Sbjct: 2 MKQGIKPIVVTYCSLMDGYCLVKEVNKAKSILYTMSQRGVN--PDIQSYNILIDGFCKIK 175
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
+V EA+ +F++M + P+V TYNSLI LCK+ ++ +LV++M + P+ +T
Sbjct: 176 KVDEAMNLFKEMHHKHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDR--GVPPDIIT 349
Query: 367 YSCLLNSLKGPEEVP---GVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEME 423
YS +L++L +V +L +++ G + Y +++ K + ++++
Sbjct: 350 YSSILDALCKNHQVDKAIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLL 529
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVI 473
G +YT+MIHG G +A+ +M + + T +++I
Sbjct: 530 VKGYNITVNTYTVMIHGFCNKGLFDEALTLLSKMKDNSCIPDAVTYEIII 679
Score = 61.2 bits (147), Expect = 9e-10
Identities = 41/160 (25%), Positives = 79/160 (48%)
Frame = +2
Query: 118 NEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQ 177
N ++D L K+ + ++ DEM R + T+S++L H+V++AI++ +
Sbjct: 248 NSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSILDALCKNHQVDKAIALLTKLKD 427
Query: 178 FGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAH 237
G+ ++ + L+ LC+ +EDA +F + + + T+ V+++G+C G
Sbjct: 428 QGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVK-GYNITVNTYTVMIHGFCNKGLFD 604
Query: 238 EAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKL 277
EA + + + C PD TY I++L K + A KL
Sbjct: 605 EALTLLSKMKDNSCIPDAVTYEIIIRSLFDKDENDKAEKL 724
>AW688886 similar to GP|3258568|gb|A Unknown protein {Arabidopsis thaliana},
partial (11%)
Length = 660
Score = 97.4 bits (241), Expect = 1e-20
Identities = 56/166 (33%), Positives = 95/166 (56%), Gaps = 4/166 (2%)
Frame = +2
Query: 243 WKDIMASKCRPDLFTYAT-FIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDA 301
+K++ + C PD+ TY T + + GK+ A L GM + + PDVV +I
Sbjct: 11 FKEMTSFDCDPDVVTYNTALLMVCVEXGKIKVAHNLVNGMSKKCKDLSPDVVTYTTLIRG 190
Query: 302 LCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCM 361
C K+ V EAL++ ++M RG +PN+ TYN+LIK LC+ ++ +K+ E++E M+ GS +
Sbjct: 191 YCRKQEVDEALDILEEMNGRGLKPNIVTYNTLIKGLCEAQKWDKMKEILEQMKGDGGS-I 367
Query: 362 PNAVTYSCLLNS---LKGPEEVPGVLERMERNGCSLSDDIYNLVLR 404
P+A T++ L+NS +E V E M++ S Y++++R
Sbjct: 368 PDACTFNTLINSHCCAGNLDEAFKVFENMKKLEVSADSASYSVLIR 505
>BF003426 similar to PIR|F96760|F96 hypothetical protein T9L24.39 [imported]
- Arabidopsis thaliana, partial (43%)
Length = 774
Score = 97.4 bits (241), Expect = 1e-20
Identities = 51/148 (34%), Positives = 82/148 (54%), Gaps = 3/148 (2%)
Frame = +2
Query: 318 MKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL--- 374
M GC P+V TY +I+ +C ++++ Y+ +E+M +K P+ VT++C L L
Sbjct: 20 MISSGCLPDVTTYKDIIEGMCLCGKIDEAYKFLEEMGKK--GYPPDIVTHNCFLKVLCHN 193
Query: 375 KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSY 434
K EE + RM C S YN+++ ++ K D+ DG +TW EME+ GC PD +Y
Sbjct: 194 KKSEEALKLYGRMIELSCIPSVQTYNMLISMFFKMDDPDGAFETWHEMEKRGCRPDTDTY 373
Query: 435 TIMIHGHYENGKMKDAMRYFREMTSKGM 462
+MI G + K +DA E+ +KG+
Sbjct: 374 GVMIEGLFNCNKAEDACILLEEVINKGI 457
Score = 82.8 bits (203), Expect = 3e-16
Identities = 55/194 (28%), Positives = 90/194 (46%), Gaps = 3/194 (1%)
Frame = +2
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
+++S C PD+ TY I+ + GK+ A K M +G PD+V NC + LC
Sbjct: 20 MISSGCLPDVTTYKDIIEGMCLCGKIDEAYKFLEEMGKKGY--PPDIVTHNCFLKVLCHN 193
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAV 365
K+ EAL+++ M E C P+V TYN LI K+ + +E +ME++ C P+
Sbjct: 194 KKSEEALKLYGRMIELSCIPSVQTYNMLISMFFKMDDPDGAFETWHEMEKR--GCRPDTD 367
Query: 366 TYSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEM 422
TY ++ L E+ +LE + G L ++ +L + N + K D M
Sbjct: 368 TYGVMIEGLFNCNKAEDACILLEEVINKGIKLPYRKFDSLLMQLSEIGNLQAIHKLSDHM 547
Query: 423 ERNGCGPDRRSYTI 436
+ P R + +
Sbjct: 548 RKFYNNPMARRFAL 589
Score = 67.0 bits (162), Expect = 2e-11
Identities = 43/139 (30%), Positives = 71/139 (50%), Gaps = 1/139 (0%)
Frame = +2
Query: 219 DIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLF 278
D+ T+ I+ G C+ G EA + +++ PD+ T+ F+K L K ALKL+
Sbjct: 44 DVTTYKDIIEGMCLCGKIDEAYKFLEEMGKKGYPPDIVTHNCFLKVLCHNKKSEEALKLY 223
Query: 279 RGMWNEGCNCKPDVVICNCIIDALCFKKRVPE-ALEVFQDMKERGCEPNVATYNSLIKHL 337
M +C P V N +I ++ FK P+ A E + +M++RGC P+ TY +I+ L
Sbjct: 224 GRMIE--LSCIPSVQTYNMLI-SMFFKMDDPDGAFETWHEMEKRGCRPDTDTYGVMIEGL 394
Query: 338 CKIRRMEKVYELVEDMERK 356
+ E L+E++ K
Sbjct: 395 FNCNKAEDACILLEEVINK 451
Score = 46.2 bits (108), Expect = 3e-05
Identities = 31/135 (22%), Positives = 56/135 (40%)
Frame = +2
Query: 151 TFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSK 210
T+ ++ K++EA + G D+ L LC K E+A L+ +
Sbjct: 53 TYKDIIEGMCLCGKIDEAYKFLEEMGKKGYPPDIVTHNCFLKVLCHNKKSEEALKLY-GR 229
Query: 211 AREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGK 270
E ++T+N++++ + + + A W ++ CRPD TY I+ L K
Sbjct: 230 MIELSCIPSVQTYNMLISMFFKMDDPDGAFETWHEMEKRGCRPDTDTYGVMIEGLFNCNK 409
Query: 271 LGTALKLFRGMWNEG 285
A L + N+G
Sbjct: 410 AEDACILLEEVINKG 454
>BQ148309 weakly similar to GP|6721172|gb| hypothetical protein {Arabidopsis
thaliana}, partial (30%)
Length = 654
Score = 93.2 bits (230), Expect = 2e-19
Identities = 44/157 (28%), Positives = 92/157 (58%)
Frame = +1
Query: 75 GFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGKMSRFEELH 134
G L+++L+ +L R++ DWK AL F WA ++ + + + ++DILG+M +++
Sbjct: 178 GIHLSENLINRLLFRYKDDWKSALAIFRWAGSHSNFKHSQQSYDMMVDILGRMKAMDKMR 357
Query: 135 QVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWL 194
++ +EM +E L+ +T + ++RRF A + ++A+ +F + GL+ + ++ LL L
Sbjct: 358 EILEEM-RQESLITLNTIAKVMRRFVGARQWKDAVRIFDDLQFLGLEKNTESMNVLLDTL 534
Query: 195 CRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWC 231
C+ K VE A ++ + + + T+N++++GWC
Sbjct: 535 CKEKFVEQAREIY--LELKHYIAPNAHTFNILIHGWC 639
Score = 38.1 bits (87), Expect = 0.008
Identities = 19/48 (39%), Positives = 29/48 (59%)
Frame = +1
Query: 296 NCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRM 343
N ++D LC +K V +A E++ ++K PN T+N LI C IRR+
Sbjct: 514 NVLLDTLCKEKFVEQAREIYLELKHY-IAPNAHTFNILIHGWCNIRRV 654
>AW774457 weakly similar to GP|10176973|dbj
gb|AAF19552.1~gene_id:MTG13.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (13%)
Length = 650
Score = 92.0 bits (227), Expect = 5e-19
Identities = 58/204 (28%), Positives = 102/204 (49%), Gaps = 3/204 (1%)
Frame = -1
Query: 226 ILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEG 285
++NG+C + EA ++ D+ PD TY + I L K G++ A +L M + G
Sbjct: 650 MINGFCKIKMVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNG 471
Query: 286 CNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEK 345
++ NC+IDALC V +A+ + + +K++G +P++ T+N LI LCK+ R++
Sbjct: 470 QPA--NIFTYNCLIDALCKNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKN 297
Query: 346 VYELVEDMERKKGSCMPNAVTYSCLLNSLKGP---EEVPGVLERMERNGCSLSDDIYNLV 402
++ +D+ K S NA TY+ ++N L +E +L +M+ NG Y +
Sbjct: 296 AQDVFQDLLSKGYSV--NAWTYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTYETL 123
Query: 403 LRLYMKWDNQDGLRKTWDEMERNG 426
++ D + K EM G
Sbjct: 122 IQALFHKDENEKAEKLLREMIARG 51
Score = 64.3 bits (155), Expect = 1e-10
Identities = 45/197 (22%), Positives = 93/197 (46%)
Frame = -1
Query: 126 KMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLD 185
K+ +E +F++M + ++ T+++L+ + ++ A + G ++
Sbjct: 632 KIKMVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNGQPANIF 453
Query: 186 AFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKD 245
+ L+ LC+ HV+ A L K ++ + D+ T+N+++ G C +G A+ V++D
Sbjct: 452 TYNCLIDALCKNHHVDQAIALV-KKIKDQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQD 276
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
+++ + +TY + L K+G A L M + G PD V +I AL K
Sbjct: 275 LLSKGYSVNAWTYNIMVNGLCKEGLFDEAEALLSKMDDNG--IIPDAVTYETLIQALFHK 102
Query: 306 KRVPEALEVFQDMKERG 322
+A ++ ++M RG
Sbjct: 101 DENEKAEKLLREMIARG 51
>BE998743 similar to GP|22128591|gb| fertility restorer-like protein {Petunia
x hybrida}, partial (7%)
Length = 829
Score = 84.3 bits (207), Expect = 1e-16
Identities = 58/211 (27%), Positives = 102/211 (47%), Gaps = 3/211 (1%)
Frame = +2
Query: 219 DIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLF 278
D+ T+N +++G+C++ ++AK V I PD +Y + + + A KL
Sbjct: 173 DVVTYNSLMDGYCLVNEVNKAKHVLSTIARMGVAPDAQSYNIMVNGFCR---ISHAWKLV 343
Query: 279 RGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLC 338
M G PD+ + +IDALC + +A+ + + +K++G +PN+ TYN LI LC
Sbjct: 344 DEMHVNGQ--PPDIFTYSSLIDALCKNNHLDKAIALVKKIKDQGIQPNMYTYNILIDGLC 517
Query: 339 KIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGP---EEVPGVLERMERNGCSLS 395
K R++ ++ +D+ K S N TY+ L+N L ++ +L +ME N + +
Sbjct: 518 KGGRLKNAQDVFQDLLTKGYSL--NIRTYNILINGLCKEGLFDKAEALLSKMEDNDINPN 691
Query: 396 DDIYNLVLRLYMKWDNQDGLRKTWDEMERNG 426
Y ++R D + K EM G
Sbjct: 692 VVTYETIIRSLFYKDYNEKAEKLLREMVARG 784
Score = 61.6 bits (148), Expect = 7e-10
Identities = 53/212 (25%), Positives = 87/212 (41%), Gaps = 35/212 (16%)
Frame = +2
Query: 289 KPDVVICNCIIDALCFKK--------------------------------RVPEALEVFQ 316
KPDVV N ++D C R+ A ++
Sbjct: 167 KPDVVTYNSLMDGYCLVNEVNKAKHVLSTIARMGVAPDAQSYNIMVNGFCRISHAWKLVD 346
Query: 317 DMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL-K 375
+M G P++ TY+SLI LCK ++K LV+ + K PN TY+ L++ L K
Sbjct: 347 EMHVNGQPPDIFTYSSLIDALCKNNHLDKAIALVKKI--KDQGIQPNMYTYNILIDGLCK 520
Query: 376 GP--EEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRS 433
G + V + + G SL+ YN+++ K D +ME N P+ +
Sbjct: 521 GGRLKNAQDVFQDLLTKGYSLNIRTYNILINGLCKEGLFDKAEALLSKMEDNDINPNVVT 700
Query: 434 YTIMIHGHYENGKMKDAMRYFREMTSKGMVAE 465
Y +I + + A + REM ++G++ E
Sbjct: 701 YETIIRSLFYKDYNEKAEKLLREMVARGLL*E 796
Score = 54.7 bits (130), Expect = 8e-08
Identities = 37/140 (26%), Positives = 71/140 (50%)
Frame = +2
Query: 183 DLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRV 242
D+ + +L+ LC+ H++ A L K ++ + ++ T+N++++G C G A+ V
Sbjct: 374 DIFTYSSLIDALCKNNHLDKAIALV-KKIKDQGIQPNMYTYNILIDGLCKGGRLKNAQDV 550
Query: 243 WKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDAL 302
++D++ ++ TY I L K+G A L M + N P+VV II +L
Sbjct: 551 FQDLLTKGYSLNIRTYNILINGLCKEGLFDKAEALLSKMEDNDIN--PNVVTYETIIRSL 724
Query: 303 CFKKRVPEALEVFQDMKERG 322
+K +A ++ ++M RG
Sbjct: 725 FYKDYNEKAEKLLREMVARG 784
Score = 39.7 bits (91), Expect = 0.003
Identities = 31/123 (25%), Positives = 57/123 (46%), Gaps = 6/123 (4%)
Frame = +2
Query: 357 KGSCMPNAVTYS------CLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWD 410
KGS P+ VTY+ CL+N + + V + RM G + YN+++ + +
Sbjct: 155 KGSIKPDVVTYNSLMDGYCLVNEVNKAKHVLSTIARM---GVAPDAQSYNIMVNGFCRIS 325
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ K DEM NG PD +Y+ +I +N + A+ +++ +G+ T
Sbjct: 326 H---AWKLVDEMHVNGQPPDIFTYSSLIDALCKNNHLDKAIALVKKIKDQGIQPNMYTYN 496
Query: 471 LVI 473
++I
Sbjct: 497 ILI 505
>TC83956 weakly similar to GP|15982931|gb|AAL09812.1 AT3g53700/F4P12_400
{Arabidopsis thaliana}, partial (18%)
Length = 762
Score = 83.6 bits (205), Expect = 2e-16
Identities = 54/217 (24%), Positives = 101/217 (45%)
Frame = +1
Query: 124 LGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLD 183
L + S F+ + + ++ + N TF+TL++ F H++E + + + G D
Sbjct: 118 LTQSSSFDSITTLLKQLKSSGSIPNATTFATLIQSFTNFHEIENLLKIL--ENELGFKPD 291
Query: 184 LDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVW 243
+ + L L ++ E L HSK + D+ T+NV++ C A +
Sbjct: 292 TNFYNIALNALVEDNKLKLVEML-HSKMVNEGIVLDVSTFNVLIKALCKAHQLRPAILML 468
Query: 244 KDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALC 303
+++ +PD T+ T ++ ++G L ALK+ + M GC V +++ C
Sbjct: 469 EEMANHGLKPDEITFTTLMQGFIEEGDLNGALKMKKQMLGYGCLLTN--VSVKVLVNGFC 642
Query: 304 FKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKI 340
+ RV EAL ++ E G P+ T+NSL+ +C+I
Sbjct: 643 KEGRVEEALRFVLEVSEEGFSPDQGTFNSLVNGVCRI 753
Score = 58.2 bits (139), Expect = 7e-09
Identities = 46/200 (23%), Positives = 92/200 (46%), Gaps = 3/200 (1%)
Frame = +1
Query: 244 KDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALC 303
K + +S P+ T+AT I++ T ++ LK+ NE KPD N ++AL
Sbjct: 160 KQLKSSGSIPNATTFATLIQSFTNFHEIENLLKILE---NE-LGFKPDTNFYNIALNALV 327
Query: 304 FKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPN 363
++ + M G +V+T+N LIK LCK ++ ++E+M P+
Sbjct: 328 EDNKLKLVEMLHSKMVNEGIVLDVSTFNVLIKALCKAHQLRPAILMLEEMANH--GLKPD 501
Query: 364 AVTYSCLLNSLKGPEEVPGVL---ERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWD 420
+T++ L+ ++ G L ++M GC L++ +++ + K + +
Sbjct: 502 EITFTTLMQGFIEEGDLNGALKMKKQMLGYGCLLTNVSVKVLVNGFCKEGRVEEALRFVL 681
Query: 421 EMERNGCGPDRRSYTIMIHG 440
E+ G PD+ ++ +++G
Sbjct: 682 EVSEEGFSPDQGTFNSLVNG 741
>TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Arabidopsis
thaliana}, partial (7%)
Length = 978
Score = 81.6 bits (200), Expect = 6e-16
Identities = 64/249 (25%), Positives = 112/249 (44%), Gaps = 4/249 (1%)
Frame = +1
Query: 130 FEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRT 189
F++ V+ M R N FS +L + E + MF ++ GL +D A+
Sbjct: 115 FDKALAVYKSMISRGIKTNCVIFSCILHCLDEMGRALEVVDMFEEFKESGLFIDRKAYNI 294
Query: 190 LLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMAS 249
L LC+ V+DA + + + QL D+K + ++NG+ + G EA+ ++K++
Sbjct: 295 LFDALCKLGKVDDAVGML-DELKSMQLDVDMKHYTTLINGYFLQGKPIEAQSLFKEMEER 471
Query: 250 KCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVP 309
+PD+ Y + A+ L M ++G +P+ II+ LC +V
Sbjct: 472 GFKPDVVAYNVLAAGFFRNRTDFEAMDLLNYMESQG--VEPNSTTHKIIIEGLCSAGKVE 645
Query: 310 EALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDM----ERKKGSCMPNAV 365
EA E F +K E +V Y +L+ C+ +EK +EL E + + P+ V
Sbjct: 646 EAEEFFNWLKGESVEISVEIYTALVNGYCEAALIEKSHELKEAFILLRTMLEMNMKPSKV 825
Query: 366 TYSCLLNSL 374
YS + +L
Sbjct: 826 MYSKIFTAL 852
Score = 69.3 bits (168), Expect = 3e-12
Identities = 52/220 (23%), Positives = 100/220 (44%), Gaps = 3/220 (1%)
Frame = +1
Query: 238 EAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNC 297
EA+ V+ ++ PD++ Y + AL +++ M + G K + VI +C
Sbjct: 16 EAESVFLEMEKQGLVPDVYVYCALVHGYCNSRNFDKALAVYKSMISRGI--KTNCVIFSC 189
Query: 298 IIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKK 357
I+ L R E +++F++ KE G + YN L LCK+ +++ +++++ K
Sbjct: 190 ILHCLDEMGRALEVVDMFEEFKESGLFIDRKAYNILFDALCKLGKVDDAVGMLDEL--KS 363
Query: 358 GSCMPNAVTYSCLLNS--LKG-PEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDG 414
+ Y+ L+N L+G P E + + ME G YN++ + +
Sbjct: 364 MQLDVDMKHYTTLINGYFLQGKPIEAQSLFKEMEERGFKPDVVAYNVLAAGFFRNRTDFE 543
Query: 415 LRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYF 454
+ ME G P+ ++ I+I G GK+++A +F
Sbjct: 544 AMDLLNYMESQGVEPNSTTHKIIIEGLCSAGKVEEAEEFF 663
Score = 66.2 bits (160), Expect = 3e-11
Identities = 42/158 (26%), Positives = 74/158 (46%), Gaps = 3/158 (1%)
Frame = +1
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
++ EA VF +M+++G P+V Y +L+ C R +K + + M + N V
Sbjct: 7 KLDEAESVFLEMEKQGLVPDVYVYCALVHGYCNSRNFDKALAVYKSMISR--GIKTNCVI 180
Query: 367 YSCLLNSLKGPE---EVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEME 423
+SC+L+ L EV + E + +G + YN++ K D DE++
Sbjct: 181 FSCILHCLDEMGRALEVVDMFEEFKESGLFIDRKAYNILFDALCKLGKVDDAVGMLDELK 360
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKG 461
D + YT +I+G++ GK +A F+EM +G
Sbjct: 361 SMQLDVDMKHYTTLINGYFLQGKPIEAQSLFKEMEERG 474
Score = 32.3 bits (72), Expect = 0.43
Identities = 13/42 (30%), Positives = 23/42 (53%)
Frame = +1
Query: 421 EMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
EME+ G PD Y ++HG+ + A+ ++ M S+G+
Sbjct: 37 EMEKQGLVPDVYVYCALVHGYCNSRNFDKALAVYKSMISRGI 162
>TC83856 weakly similar to PIR|B96656|B96656 unknown protein 41955-40111
[imported] - Arabidopsis thaliana, partial (12%)
Length = 662
Score = 80.5 bits (197), Expect = 1e-15
Identities = 54/164 (32%), Positives = 83/164 (49%), Gaps = 4/164 (2%)
Frame = +2
Query: 252 RPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEA 311
+PD++T+ + K GK+ ALKL M + G P++V + I+DALC RV +A
Sbjct: 71 KPDVYTFNILVDVFCKSGKISYALKLVDEMHDRG--QPPNIVTYSSILDALCKTHRVDKA 244
Query: 312 LEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLL 371
+ + +K++G PN+ TY LI LC ++E + ED+ K VTY +
Sbjct: 245 VALLTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDI--TVVTYIVMF 418
Query: 372 NSL--KGP-EEVPGVLERMERNGCSLSDDIYNLV-LRLYMKWDN 411
KG +E +L +ME NGC Y L+ L L+ K +N
Sbjct: 419 YGFCKKGLFDEASALLSKMEENGCIPDAKTYELIKLSLFKKGEN 550
Score = 78.2 bits (191), Expect = 7e-15
Identities = 46/163 (28%), Positives = 81/163 (49%), Gaps = 3/163 (1%)
Frame = +2
Query: 287 NCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKV 346
N KPDV N ++D C ++ AL++ +M +RG PN+ TY+S++ LCK R++K
Sbjct: 65 NIKPDVYTFNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDKA 244
Query: 347 YELVEDMERKKGSCMPNAVTYSCLLNSLKGP---EEVPGVLERMERNGCSLSDDIYNLVL 403
L+ + K PN TY+ L++ L E+ + E + G ++ Y ++
Sbjct: 245 VALLTKL--KDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDITVVTYIVMF 418
Query: 404 RLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGK 446
+ K D +ME NGC PD ++Y ++ ++ G+
Sbjct: 419 YGFCKKGLFDEASALLSKMEENGCIPDAKTYELIKLSLFKKGE 547
Score = 59.3 bits (142), Expect = 3e-09
Identities = 42/171 (24%), Positives = 79/171 (45%), Gaps = 1/171 (0%)
Frame = +2
Query: 108 DSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEE 167
++ P N ++D+ K + ++ DEM R N T+S++L H+V++
Sbjct: 62 ENIKPDVYTFNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDK 241
Query: 168 AISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHS-KAREFQLHRDIKTWNVI 226
A+++ + G+ ++ + L+ LC +EDA +F + + + + T+ V+
Sbjct: 242 AVALLTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDI--TVVTYIVM 415
Query: 227 LNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKL 277
G+C G EA + + + C PD TY +L KKG+ A KL
Sbjct: 416 FYGFCKKGLFDEASALLSKMEENGCIPDAKTYELIKLSLFKKGENDMAEKL 568
Score = 51.6 bits (122), Expect = 7e-07
Identities = 30/136 (22%), Positives = 62/136 (45%)
Frame = +2
Query: 151 TFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSK 210
TF+ L+ F + K+ A+ + G ++ + ++L LC+ V+ A L +K
Sbjct: 86 TFNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDKAVALL-TK 262
Query: 211 AREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGK 270
++ + ++ T+ ++++G C G +A+ +++D++ + TY KKG
Sbjct: 263 LKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDITVVTYIVMFYGFCKKGL 442
Query: 271 LGTALKLFRGMWNEGC 286
A L M GC
Sbjct: 443 FDEASALLSKMEENGC 490
Score = 45.4 bits (106), Expect = 5e-05
Identities = 33/138 (23%), Positives = 59/138 (41%)
Frame = +2
Query: 324 EPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGV 383
+P+V T+N L+ CK ++ +LV++M + PN VTYS +L++L V
Sbjct: 71 KPDVYTFNILVDVFCKSGKISYALKLVDEMHDRGQP--PNIVTYSSILDALCKTHRVDKA 244
Query: 384 LERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYE 443
+ + + ++ G P+ +YTI+I G
Sbjct: 245 VALLTK--------------------------------LKDQGIRPNMHTYTILIDGLCT 328
Query: 444 NGKMKDAMRYFREMTSKG 461
+GK++DA F ++ KG
Sbjct: 329 SGKLEDARNIFEDLLVKG 382
>TC82855 weakly similar to PIR|T45829|T45829 hypothetical protein F2K15.100
- Arabidopsis thaliana, partial (63%)
Length = 1447
Score = 68.6 bits (166), Expect = 5e-12
Identities = 41/152 (26%), Positives = 76/152 (49%), Gaps = 3/152 (1%)
Frame = +3
Query: 226 ILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGM---W 282
++ G+ + G EA ++++ + Y + + AL K GK A++LF M
Sbjct: 402 LMKGYFLKGMEKEAMECYQEVFVEGKKMSDIAYNSVLDALAKNGKFDEAMRLFDRMIKEH 581
Query: 283 NEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRR 342
N ++ N ++D C + + EA+EVF+ M E C+P+ ++N+LI+ LC
Sbjct: 582 NPPAKLAVNLGSFNVMVDGYCAEGKFKEAIEVFRSMGESRCKPDTLSFNNLIEQLCNNGM 761
Query: 343 MEKVYELVEDMERKKGSCMPNAVTYSCLLNSL 374
+ + E+ +ME K P+ TY L+++L
Sbjct: 762 ILEAEEVYGEMEGK--GVNPDEYTYGLLMDTL 851
Score = 56.2 bits (134), Expect(2) = 2e-15
Identities = 46/202 (22%), Positives = 96/202 (46%), Gaps = 6/202 (2%)
Frame = +3
Query: 107 ADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNED--TFSTLLRRFAAAHK 164
A +AP V + ++ + S + + +V++E+ + G V ED L++ +
Sbjct: 252 ASGFAPDPLVYHYLMLGHARSSDGDGVLRVYEELKEKLGGVVEDGVVLGCLMKGYFLKGM 431
Query: 165 VEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREF----QLHRDI 220
+EA+ + G + A+ ++L L + ++A LF +E +L ++
Sbjct: 432 EKEAMECYQEVFVEGKKMSDIAYNSVLDALAKNGKFDEAMRLFDRMIKEHNPPAKLAVNL 611
Query: 221 KTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRG 280
++NV+++G+C G EA V++ + S+C+PD ++ I+ L G + A +++
Sbjct: 612 GSFNVMVDGYCAEGKFKEAIEVFRSMGESRCKPDTLSFNNLIEQLCNNGMILEAEEVYGE 791
Query: 281 MWNEGCNCKPDVVICNCIIDAL 302
M +G N PD ++D L
Sbjct: 792 MEGKGVN--PDEYTYGLLMDTL 851
Score = 43.9 bits (102), Expect(2) = 2e-15
Identities = 32/127 (25%), Positives = 56/127 (43%), Gaps = 5/127 (3%)
Frame = +1
Query: 303 CFKKRVPE-ALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCM 361
CFK+ P+ A F+ M E G PN+A YN L+ L K+ +++ + M +K
Sbjct: 850 CFKENRPDDAASYFKKMVESGLRPNLAVYNRLVDGLVKVGKIDDAKFFFDLMVKK---LK 1020
Query: 362 PNAVTYSCLLNSLKGPEEVPGVLE----RMERNGCSLSDDIYNLVLRLYMKWDNQDGLRK 417
+ +Y ++ L + VL+ ++ NG ++ V K ++ L K
Sbjct: 1021MDVASYQFMMKVLSEAGRLDDVLQIVNMLLDDNGVDFDEEFQEFVKVELRKEGREEDLAK 1200
Query: 418 TWDEMER 424
+E ER
Sbjct: 1201LMEEKER 1221
Score = 42.4 bits (98), Expect = 4e-04
Identities = 48/208 (23%), Positives = 76/208 (36%), Gaps = 3/208 (1%)
Frame = +3
Query: 253 PDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEAL 312
P++ TY + K TAL+ F+ + + P +I L R+ A+
Sbjct: 54 PNIITYNLIFQTYLDCRKPDTALENFK-QFIKDAPFNPSPTTFRILIKGLVDNNRLDRAM 230
Query: 313 EVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLN 372
++ M G P+ Y+ L+ + + V + E+++ K G + + V CL+
Sbjct: 231 DIKDQMNASGFAPDPLVYHYLMLGHARSSDGDGVLRVYEELKEKLGGVVEDGVVLGCLMK 410
Query: 373 S--LKGPE-EVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGP 429
LKG E E + + G +SD YN VL D L K
Sbjct: 411 GYFLKGMEKEAMECYQEVFVEGKKMSDIAYNSVL---------DALAK------------ 527
Query: 430 DRRSYTIMIHGHYENGKMKDAMRYFREM 457
NGK +AMR F M
Sbjct: 528 --------------NGKFDEAMRLFDRM 569
Score = 38.9 bits (89), Expect = 0.005
Identities = 40/223 (17%), Positives = 82/223 (35%), Gaps = 6/223 (2%)
Frame = +3
Query: 222 TWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGM 281
T+ +++ G A + + AS PD Y + + L+++ +
Sbjct: 174 TFRILIKGLVDNNRLDRAMDIKDQMNASGFAPDPLVYHYLMLGHARSSDGDGVLRVYEEL 353
Query: 282 WNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIR 341
+ D V+ C++ K EA+E +Q++ G + + YNS++ L K
Sbjct: 354 KEKLGGVVEDGVVLGCLMKGYFLKGMEKEAMECYQEVFVEGKKMSDIAYNSVLDALAKNG 533
Query: 342 RMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGP------EEVPGVLERMERNGCSLS 395
+ ++ L + M ++ AV + G +E V M + C
Sbjct: 534 KFDEAMRLFDRMIKEHNPPAKLAVNLGSFNVMVDGYCAEGKFKEAIEVFRSMGESRCKPD 713
Query: 396 DDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMI 438
+N ++ + + EME G PD +Y +++
Sbjct: 714 TLSFNNLIEQLCNNGMILEAEEVYGEMEGKGVNPDEYTYGLLM 842
Score = 38.1 bits (87), Expect = 0.008
Identities = 28/149 (18%), Positives = 67/149 (44%), Gaps = 5/149 (3%)
Frame = +3
Query: 318 MKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL--- 374
+ + G PN+ TYN + + R+ + E + K P+ T+ L+ L
Sbjct: 33 LTQAGVVPNIITYNLIFQTYLDCRKPDTALENFKQF-IKDAPFNPSPTTFRILIKGLVDN 209
Query: 375 KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMER--NGCGPDRR 432
+ + ++M +G + +Y+ ++ + + + DG+ + ++E++ G D
Sbjct: 210 NRLDRAMDIKDQMNASGFAPDPLVYHYLMLGHARSSDGDGVLRVYEELKEKLGGVVEDGV 389
Query: 433 SYTIMIHGHYENGKMKDAMRYFREMTSKG 461
++ G++ G K+AM ++E+ +G
Sbjct: 390 VLGCLMKGYFLKGMEKEAMECYQEVFVEG 476
Score = 37.0 bits (84), Expect = 0.017
Identities = 34/156 (21%), Positives = 59/156 (37%), Gaps = 5/156 (3%)
Frame = +3
Query: 118 NEILDILGKMSRFEELHQVFDEMSHREG-----LVNEDTFSTLLRRFAAAHKVEEAISMF 172
N +LD L K +F+E ++FD M VN +F+ ++ + A K +EAI +F
Sbjct: 501 NSVLDALAKNGKFDEAMRLFDRMIKEHNPPAKLAVNLGSFNVMVDGYCAEGKFKEAIEVF 680
Query: 173 YTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCV 232
+ + D +F L+ LC NG +
Sbjct: 681 RSMGESRCKPDTLSFNNLIEQLCN-------------------------------NGMIL 767
Query: 233 LGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKK 268
EA+ V+ ++ PD +TY + L ++
Sbjct: 768 -----EAEEVYGEMEGKGVNPDEYTYGLLMDTLFQR 860
Score = 31.6 bits (70), Expect = 0.73
Identities = 30/121 (24%), Positives = 52/121 (42%), Gaps = 1/121 (0%)
Frame = +1
Query: 238 EAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNC 297
+A +K ++ S RP+L Y + L K GK+ A F M + K DV
Sbjct: 874 DAASYFKKMVESGLRPNLAVYNRLVDGLVKVGKIDDAKFFFDLMVKK---LKMDVASYQF 1044
Query: 298 IIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIK-HLCKIRRMEKVYELVEDMERK 356
++ L R+ + L++ + + + +K L K R E + +L+E+ ER
Sbjct: 1045MMKVLSEAGRLDDVLQIVNMLLDDNGVDFDEEFQEFVKVELRKEGREEDLAKLMEEKERL 1224
Query: 357 K 357
K
Sbjct: 1225K 1227
>TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein AT4g19900
[imported] - Arabidopsis thaliana, partial (18%)
Length = 756
Score = 79.0 bits (193), Expect = 4e-15
Identities = 62/277 (22%), Positives = 115/277 (41%)
Frame = +1
Query: 187 FRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDI 246
+ ++ CR + AE L S+ +E L + T+ +++G C GN A + +
Sbjct: 1 YTAMIRGYCREDKLNRAEMLL-SRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLM 177
Query: 247 MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKK 306
+ P+L TY + L K+G++ A K+ + G KPD N ++ C ++
Sbjct: 178 SSEGFSPNLCTYNAIVNGLCKRGRVQEAYKMLEDGFQNGL--KPDKFTYNILMSEHCKQE 351
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
+ +AL +F M + G +P++ +Y +LI C+ RM++ E+ R +P T
Sbjct: 352 NIRQALALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRI--GIIPTNKT 525
Query: 367 YSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNG 426
Y+ ++ C Y + N K + + +G
Sbjct: 526 YTSMI--------------------CG------------YCREGNLTLAMKFFHRLSDHG 609
Query: 427 CGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMV 463
C P+ +Y +I G + K +A + M KG+V
Sbjct: 610 CAPESITYGAIISGLCKQSKRDEARSLYDSMIEKGLV 720
Score = 78.2 bits (191), Expect = 7e-15
Identities = 54/220 (24%), Positives = 97/220 (43%), Gaps = 3/220 (1%)
Frame = +1
Query: 258 YATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQD 317
Y I+ ++ KL A L M +G P+ +ID C A ++
Sbjct: 1 YTAMIRGYCREDKLNRAEMLLSRMKEQGL--VPNTNTYTTLIDGHCKAGNFERAYDLMNL 174
Query: 318 MKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGP 377
M G PN+ TYN+++ LCK R+++ Y+++ED + P+ TY+ L++
Sbjct: 175 MSSEGFSPNLCTYNAIVNGLCKRGRVQEAYKMLED--GFQNGLKPDKFTYNILMSEHCKQ 348
Query: 378 EEVPGVL---ERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSY 434
E + L +M + G Y ++ ++ + + ++E R G P ++Y
Sbjct: 349 ENIRQALALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRIGIIPTNKTY 528
Query: 435 TIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVIS 474
T MI G+ G + AM++F ++ G E T +IS
Sbjct: 529 TSMICGYCREGNLTLAMKFFHRLSDHGCAPESITYGAIIS 648
Score = 75.9 bits (185), Expect = 3e-14
Identities = 53/223 (23%), Positives = 100/223 (44%)
Frame = +1
Query: 152 FSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKA 211
++ ++R + K+ A + ++ GL + + + TL+ C+ + E A L + +
Sbjct: 1 YTAMIRGYCREDKLNRAEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMS 180
Query: 212 REFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKL 271
E ++ T+N I+NG C G EA ++ +D + +PD FTY + K+ +
Sbjct: 181 SE-GFSPNLCTYNAIVNGLCKRGRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQENI 357
Query: 272 GTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYN 331
AL LF M G +PD+ +I C + R+ E+ F++ G P TY
Sbjct: 358 RQALALFNKMLKIG--IQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRIGIIPTNKTYT 531
Query: 332 SLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL 374
S+I C+ + + + C P ++TY +++ L
Sbjct: 532 SMICGYCREGNLTLAMKFFHRL--SDHGCAPESITYGAIISGL 654
Score = 71.2 bits (173), Expect = 8e-13
Identities = 53/243 (21%), Positives = 102/243 (41%)
Frame = +1
Query: 106 KADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKV 165
K P + ++D K FE + + + MS N T++ ++ +V
Sbjct: 73 KEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMSSEGFSPNLCTYNAIVNGLCKRGRV 252
Query: 166 EEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNV 225
+EA M Q GL D + L+ C+ +++ A LF+ K + + DI ++
Sbjct: 253 QEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQENIRQALALFN-KMLKIGIQPDIHSYTT 429
Query: 226 ILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEG 285
++ +C E++ +++ + P TY + I ++G L A+K F + + G
Sbjct: 430 LIAVFCRENRMKESEMFFEEAVRIGIIPTNKTYTSMICGYCREGNLTLAMKFFHRLSDHG 609
Query: 286 CNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEK 345
C P+ + II LC + + EA ++ M E+G P C++ ++
Sbjct: 610 C--APESITYGAIISGLCKQSKRDEARSLYDSMIEKGLVP------------CEVTKITL 747
Query: 346 VYE 348
YE
Sbjct: 748 AYE 756
>BF004907 weakly similar to GP|8953393|emb putative protein {Arabidopsis
thaliana}, partial (34%)
Length = 665
Score = 78.2 bits (191), Expect = 7e-15
Identities = 50/209 (23%), Positives = 103/209 (48%), Gaps = 3/209 (1%)
Frame = +3
Query: 151 TFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLC---RYKHVEDAETLF 207
T+ TL+ + +VE+A+ M + G+ + + ++ L R+K F
Sbjct: 33 TYGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEALGMMERF 212
Query: 208 HSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTK 267
H Q+ + T+N ++ G+C G+ A ++ K +++ P TY F + ++
Sbjct: 213 HV----LQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFFRYFSR 380
Query: 268 KGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNV 327
GK+ + L+ M G N PD + + ++ LC ++++ A++V +M+ +G + ++
Sbjct: 381 CGKVDEGMNLYTKMIESGHN--PDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKGYDMDL 554
Query: 328 ATYNSLIKHLCKIRRMEKVYELVEDMERK 356
AT L LCK+ ++E+ + EDM R+
Sbjct: 555 ATSTMLTHLLCKMHKLEEAFAEFEDMIRR 641
Score = 67.0 bits (162), Expect = 2e-11
Identities = 45/178 (25%), Positives = 82/178 (45%), Gaps = 3/178 (1%)
Frame = +3
Query: 287 NCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKV 346
N +P VV +++ C +RV +ALE+ +M + G +PN YN +I L + R ++
Sbjct: 12 NVRPSVVTYGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEA 191
Query: 347 YELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPG---VLERMERNGCSLSDDIYNLVL 403
++E + P TY+ L+ ++ G +L++M G YN
Sbjct: 192 LGMMERFHVLQ--IGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFF 365
Query: 404 RLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKG 461
R + + D + +M +G PDR +Y +++ E K++ A++ EM KG
Sbjct: 366 RYFSRCGKVDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKG 539
Score = 28.9 bits (63), Expect = 4.7
Identities = 21/88 (23%), Positives = 34/88 (37%)
Frame = +3
Query: 125 GKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDL 184
GK+ L+ E H + T+ +L+ K+E A+ + G D+DL
Sbjct: 384 GKVDEGMNLYTKMIESGHNPDRL---TYHLVLKMLCEEEKLELAVQVSMEMRHKGYDMDL 554
Query: 185 DAFRTLLMWLCRYKHVEDAETLFHSKAR 212
L LC+ +E+A F R
Sbjct: 555 ATSTMLTHLLCKMHKLEEAFAEFEDMIR 638
>TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16.90 -
Arabidopsis thaliana, partial (43%)
Length = 1038
Score = 73.2 bits (178), Expect = 2e-13
Identities = 47/202 (23%), Positives = 101/202 (49%), Gaps = 3/202 (1%)
Frame = +1
Query: 289 KPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYE 348
+PDVV +I+A +R EAL VF++M + G P YN L+ +E+
Sbjct: 91 RPDVVSYALLINAYGKARREEEALAVFEEMLDAGVRPTRKAYNILLDAFSISGMVEQARI 270
Query: 349 LVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGV---LERMERNGCSLSDDIYNLVLRL 405
+ + M R K MP+ +Y+ +L++ ++ G +R+ ++G + Y +++
Sbjct: 271 VFKSMRRDK--YMPDLCSYTTMLSAYVNAPDMEGAEKFFKRLIQDGFEPNVVTYGTLIKG 444
Query: 406 YMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAE 465
Y K ++ + + + ++EM G ++ T ++ H +NG A+ +F+EM G++ +
Sbjct: 445 YAKANDIEKVMEKYEEMLGRGIKANQTILTTIMDAHGKNGDFDSAVNWFKEMALNGLLPD 624
Query: 466 PRTEKLVISMNSPLKERTEKQE 487
+ + +++S+ ++ E E
Sbjct: 625 QKAKNILLSLAKTEEDIKEANE 690
Score = 57.8 bits (138), Expect = 1e-08
Identities = 50/241 (20%), Positives = 102/241 (41%), Gaps = 16/241 (6%)
Frame = +1
Query: 128 SRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAF 187
+ ++E+ ++D+M + + +++ L+ + A + EEA+++F G+ A+
Sbjct: 37 TNYKEVSNIYDQMQRADLRPDVVSYALLINAYGKARREEEALAVFEEMLDAGVRPTRKAY 216
Query: 188 RTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIM 247
LL VE A +F S R+ + D+ ++ +L+ + + A++ +K ++
Sbjct: 217 NILLDAFSISGMVEQARIVFKSMRRD-KYMPDLCSYTTMLSAYVNAPDMEGAEKFFKRLI 393
Query: 248 ASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKR 307
P++ TY T IK K + ++ + M G K + I I+DA
Sbjct: 394 QDGFEPNVVTYGTLIKGYAKANDIEKVMEKYEEMLGRG--IKANQTILTTIMDAHGKNGD 567
Query: 308 VPEALEVFQDMKERG----------------CEPNVATYNSLIKHLCKIRRMEKVYELVE 351
A+ F++M G E ++ N L+ H +I + KV + E
Sbjct: 568 FDSAVNWFKEMALNGLLPDQKAKNILLSLAKTEEDIKEANELVLHSIEINNLPKVNDHEE 747
Query: 352 D 352
D
Sbjct: 748 D 750
>BF649699 similar to GP|9757911|dbj gb|AAF19552.1~gene_id:MMN10.14~similar to
unknown protein {Arabidopsis thaliana}, partial (11%)
Length = 617
Score = 72.8 bits (177), Expect = 3e-13
Identities = 49/203 (24%), Positives = 95/203 (46%)
Frame = +1
Query: 165 VEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWN 224
++ A + +FG +L + L+ LC+ + ++ AE L + L + T++
Sbjct: 22 IDSAYDLVVKLGRFGFLPNLFVYNALINALCKGEDLDKAE-LLYKNMHSMNLPLNDVTYS 198
Query: 225 VILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNE 284
++++ +C G A+ + ++ R ++ Y + I K G L A L+ M NE
Sbjct: 199 ILIDSFCKRGMLDVAESYFGRMIEDGIRETIYPYNSLINGHCKFGDLSAAEFLYTKMINE 378
Query: 285 GCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRME 344
G +P +I C +V +A +++++M E+ P+V T+ +LI LC M
Sbjct: 379 G--LEPTATTFTTLISGYCKDLQVEKAFKLYREMNEKEXAPSVYTFTALIYGLCSTNEMA 552
Query: 345 KVYELVEDMERKKGSCMPNAVTY 367
+ +L ++M +K P VTY
Sbjct: 553 EASKLFDEMVERK--IKPTXVTY 615
Score = 57.8 bits (138), Expect = 1e-08
Identities = 40/171 (23%), Positives = 79/171 (45%)
Frame = +1
Query: 210 KAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKG 269
K F ++ +N ++N C + +A+ ++K++ + + TY+ I + K+G
Sbjct: 49 KLGRFGFLPNLFVYNALINALCKGEDLDKAELLYKNMHSMNLPLNDVTYSILIDSFCKRG 228
Query: 270 KLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVAT 329
L A F M +G + + N +I+ C + A ++ M G EP T
Sbjct: 229 MLDVAESYFGRMIEDG--IRETIYPYNSLINGHCKFGDLSAAEFLYTKMINEGLEPTATT 402
Query: 330 YNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEV 380
+ +LI CK ++EK ++L +M K+ + P+ T++ L+ L E+
Sbjct: 403 FTTLISGYCKDLQVEKAFKLYREMNEKEXA--PSVYTFTALIYGLCSTNEM 549
Score = 48.9 bits (115), Expect = 4e-06
Identities = 48/201 (23%), Positives = 85/201 (41%), Gaps = 5/201 (2%)
Frame = +1
Query: 265 LTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCE 324
L KKG + +A L + G P++ + N +I+ALC + + +A ++++M
Sbjct: 4 LRKKGNIDSAYDLVVKLGRFGF--LPNLFVYNALINALCKGEDLDKAELLYKNMHSMNLP 177
Query: 325 PNVATYNSLIKHLCKIRRMEKVYE-----LVEDMERKKGSCMPNAVTYSCLLNSLKGPEE 379
N TY+ LI CK R M V E ++ED R+ + + C L E
Sbjct: 178 LNDVTYSILIDSFCK-RGMLDVAESYFGRMIEDGIRETIYPYNSLINGHCKFGDLSAAE- 351
Query: 380 VPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIH 439
+ +M G + + ++ Y K + K + EM P ++T +I+
Sbjct: 352 --FLYTKMINEGLEPTATTFTTLISGYCKDLQVEKAFKLYREMNEKEXAPSVYTFTALIY 525
Query: 440 GHYENGKMKDAMRYFREMTSK 460
G +M +A + F EM +
Sbjct: 526 GLCSTNEMAEASKLFDEMVER 588
>TC80520 weakly similar to PIR|T02562|T02562 probable salt-inducible protein
[imported] - Arabidopsis thaliana, partial (6%)
Length = 1911
Score = 72.4 bits (176), Expect = 4e-13
Identities = 49/191 (25%), Positives = 86/191 (44%), Gaps = 3/191 (1%)
Frame = +1
Query: 253 PDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEAL 312
P ++ + A K G+ KL+ M G DV C+I A C + +V
Sbjct: 463 PSASSFNVCLLACLKVGRTDLVWKLYELMIESGVGVNIDVETVGCLIKAFCAENKVFNGY 642
Query: 313 EVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLN 372
E+ + + E+G + +N+LI CK ++ ++V E++ M K C P+ TY ++N
Sbjct: 643 ELLRQVLEKGLCVDNTVFNALINGFCKQKQYDRVSEILHIMIAMK--CNPSIYTYQEIIN 816
Query: 373 SL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGP 429
L + +E V ++ G +Y V++ + RK W EM + G P
Sbjct: 817 GLLKRRKNDEAFRVFNDLKDRGYFPDRVMYTTVIKGFCDMGLLAEARKLWFEMIQKGLVP 996
Query: 430 DRRSYTIMIHG 440
+ +Y +MI+G
Sbjct: 997 NEYTYNVMIYG 1029
Score = 68.6 bits (166), Expect = 5e-12
Identities = 58/280 (20%), Positives = 110/280 (38%), Gaps = 34/280 (12%)
Frame = +1
Query: 89 RHRSDWKPALVFFNWASKADSYAPTSRVCNEILD-------------------------- 122
+H+++ +L F +W + + P CN + D
Sbjct: 193 KHQNNTFLSLRFLHWLTSHCGFKPDQSSCNALFDALVDAGAVKAAKSLLEYPDFVPKNDS 372
Query: 123 ------ILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMF--YT 174
+LG+ EE+ VF + L + +F+ L + + ++
Sbjct: 373 LEGYVRLLGENGMVEEVFDVFVSLKKVGFLPSASSFNVCLLACLKVGRTDLVWKLYELMI 552
Query: 175 REQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLG 234
G+++D++ L+ C V + L + E L D +N ++NG+C
Sbjct: 553 ESGVGVNIDVETVGCLIKAFCAENKVFNGYELLR-QVLEKGLCVDNTVFNALINGFCKQK 729
Query: 235 NAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVI 294
+ ++A KC P ++TY I L K+ K A ++F + + G PD V+
Sbjct: 730 QYDRVSEILHIMIAMKCNPSIYTYQEIINGLLKRRKNDEAFRVFNDLKDRG--YFPDRVM 903
Query: 295 CNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLI 334
+I C + EA +++ +M ++G PN TYN +I
Sbjct: 904 YTTVIKGFCDMGLLAEARKLWFEMIQKGLVPNEYTYNVMI 1023
Score = 63.5 bits (153), Expect = 2e-10
Identities = 40/147 (27%), Positives = 73/147 (49%)
Frame = +3
Query: 230 WCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCK 289
+C + + EA++++ D+ ++ +Y+T I L GK AL LF M +G
Sbjct: 1029 YCKVRDFAEARKLYDDMCGRGYAENVVSYSTMISGLYLHGKTDEALSLFHEMSRKGI--A 1202
Query: 290 PDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYEL 349
D++ N +I LC + + +A + + +G EP+V+++ LIK LCK+ E L
Sbjct: 1203 RDLISYNSLIKGLCQEGELAKATNLLNKLLIQGLEPSVSSFTPLIKCLCKVGDTEGAMRL 1382
Query: 350 VEDMERKKGSCMPNAVTYSCLLNSLKG 376
++DM + + + Y + S KG
Sbjct: 1383 LKDMHDRHLEPIASTHDYMIIGLSKKG 1463
Score = 54.3 bits (129), Expect = 1e-07
Identities = 76/341 (22%), Positives = 127/341 (36%), Gaps = 74/341 (21%)
Frame = +1
Query: 207 FHSKAREFQLHRDIKTWNVI--LNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKA 264
F + E +D K N LN C + + K W++ + S+ F+ F +
Sbjct: 16 FRNLTTETTTKQDPKDQNFTRTLNEICTITRS---KPRWENTLISQYPSFNFSNPKFFLS 186
Query: 265 LTKKGKLGTALKLFRGMW-NEGCNCKPDVVICNCIIDALC-------------FKKRVP- 309
K T L L W C KPD CN + DAL + VP
Sbjct: 187 YLKHQN-NTFLSLRFLHWLTSHCGFKPDQSSCNALFDALVDAGAVKAAKSLLEYPDFVPK 363
Query: 310 ------------------EALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVE 351
E +VF +K+ G P+ +++N + K+ R + V++L E
Sbjct: 364 NDSLEGYVRLLGENGMVEEVFDVFVSLKKVGFLPSASSFNVCLLACLKVGRTDLVWKLYE 543
Query: 352 -----------DMER-------------------------KKGSCMPNAVTYSCLLNSL- 374
D+E +KG C+ N V ++ L+N
Sbjct: 544 LMIESGVGVNIDVETVGCLIKAFCAENKVFNGYELLRQVLEKGLCVDNTV-FNALINGFC 720
Query: 375 --KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRR 432
K + V +L M C+ S Y ++ +K D + +++++ G PDR
Sbjct: 721 KQKQYDRVSEILHIMIAMKCNPSIYTYQEIINGLLKRRKNDEAFRVFNDLKDRGYFPDRV 900
Query: 433 SYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVI 473
YT +I G + G + +A + + EM KG+V T ++I
Sbjct: 901 MYTTVIKGFCDMGLLAEARKLWFEMIQKGLVPNEYTYNVMI 1023
Score = 52.8 bits (125), Expect = 3e-07
Identities = 38/171 (22%), Positives = 82/171 (47%)
Frame = +3
Query: 186 AFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKD 245
++ T++ L + ++A +LFH +R+ + RD+ ++N ++ G C G +A +
Sbjct: 1110 SYSTMISGLYLHGKTDEALSLFHEMSRK-GIARDLISYNSLIKGLCQEGELAKATNLLNK 1286
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
++ P + ++ IK L K G A++L + M + + +P + +I L K
Sbjct: 1287 LLIQGLEPSVSSFTPLIKCLCKVGDTEGAMRLLKDMHDR--HLEPIASTHDYMIIGLSKK 1460
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERK 356
+ +E M +PN+ T+ LI L + R++ + +++ M R+
Sbjct: 1461 GDFAQGMEWLLKMLSWKLKPNMQTFEHLIDCLKRENRLDDILIVLDLMFRE 1613
Score = 39.7 bits (91), Expect = 0.003
Identities = 39/198 (19%), Positives = 78/198 (38%), Gaps = 36/198 (18%)
Frame = +1
Query: 106 KADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNED--TFSTLLRRFAAAH 163
K + P++ N L K+ R + + ++++ M VN D T L++ F A +
Sbjct: 445 KKVGFLPSASSFNVCLLACLKVGRTDLVWKLYELMIESGVGVNIDVETVGCLIKAFCAEN 624
Query: 164 KVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHS-------------- 209
KV + + GL +D F L+ C+ K + + H
Sbjct: 625 KVFNGYELLRQVLEKGLCVDNTVFNALINGFCKQKQYDRVSEILHIMIAMKCNPSIYTYQ 804
Query: 210 -------KARE----FQLHRDIKT---------WNVILNGWCVLGNAHEAKRVWKDIMAS 249
K R+ F++ D+K + ++ G+C +G EA+++W +++
Sbjct: 805 EIINGLLKRRKNDEAFRVFNDLKDRGYFPDRVMYTTVIKGFCDMGLLAEARKLWFEMIQK 984
Query: 250 KCRPDLFTYATFIKALTK 267
P+ +TY I + +
Sbjct: 985 GLVPNEYTYNVMIYGIVR 1038
>BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01051 protein
domain and multiple PPR PF|01535 repeats. EST
gb|AA728420 comes, partial (13%)
Length = 508
Score = 71.2 bits (173), Expect = 8e-13
Identities = 37/132 (28%), Positives = 72/132 (54%)
Frame = +1
Query: 221 KTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRG 280
+++++++NG+C + +A ++ ++ + PD TY + I L K G++ A +L
Sbjct: 118 RSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAWELVDE 297
Query: 281 MWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKI 340
M + G D++ N +IDALC V +A+ + + +K++G + ++ TYN LI LCK
Sbjct: 298 MRDSGQPA--DIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILIDGLCKQ 471
Query: 341 RRMEKVYELVED 352
R+ + D
Sbjct: 472 GRLNDAQVIFHD 507
Score = 59.3 bits (142), Expect = 3e-09
Identities = 41/132 (31%), Positives = 63/132 (47%), Gaps = 3/132 (2%)
Frame = +1
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCK---PDVVICNCIIDAL 302
I + P+ +Y+ I K + AL LF M C+ PD V N +ID L
Sbjct: 88 ISRMRVAPNARSYSIVINGFCKIKMVDKALSLFYEM-----RCRRIAPDTVTYNSLIDGL 252
Query: 303 CFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMP 362
C R+ A E+ +M++ G ++ TYNSLI LCK ++K LV+ + K
Sbjct: 253 CKSGRISYAWELVDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKI--KDQGIQL 426
Query: 363 NAVTYSCLLNSL 374
+ TY+ L++ L
Sbjct: 427 DMYTYNILIDGL 462
Score = 51.6 bits (122), Expect = 7e-07
Identities = 35/132 (26%), Positives = 67/132 (50%)
Frame = +1
Query: 186 AFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKD 245
++ ++ C+ K V+ A +LF+ + R ++ D T+N +++G C G A + +
Sbjct: 121 SYSIVINGFCKIKMVDKALSLFY-EMRCRRIAPDTVTYNSLIDGLCKSGRISYAWELVDE 297
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
+ S D+ TY + I AL K + A+ L + + ++G D+ N +ID LC +
Sbjct: 298 MRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQL--DMYTYNILIDGLCKQ 471
Query: 306 KRVPEALEVFQD 317
R+ +A +F D
Sbjct: 472 GRLNDAQVIFHD 507
Score = 48.1 bits (113), Expect = 8e-06
Identities = 26/98 (26%), Positives = 49/98 (49%)
Frame = +1
Query: 111 APTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAIS 170
AP + N ++D L K R ++ DEM + T+++L+ H V++AI+
Sbjct: 211 APDTVTYNSLIDGLCKSGRISYAWELVDEMRDSGQPADIITYNSLIDALCKNHHVDKAIA 390
Query: 171 MFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFH 208
+ + G+ LD+ + L+ LC+ + DA+ +FH
Sbjct: 391 LVKKIKDQGIQLDMYTYNILIDGLCKQGRLNDAQVIFH 504
Score = 47.8 bits (112), Expect = 1e-05
Identities = 33/131 (25%), Positives = 62/131 (47%)
Frame = +1
Query: 148 NEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLF 207
N ++S ++ F V++A+S+FY + D + +L+ LC+ + A L
Sbjct: 112 NARSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAWELV 291
Query: 208 HSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTK 267
+ R+ DI T+N +++ C + +A + K I + D++TY I L K
Sbjct: 292 -DEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILIDGLCK 468
Query: 268 KGKLGTALKLF 278
+G+L A +F
Sbjct: 469 QGRLNDAQVIF 501
Score = 47.4 bits (111), Expect = 1e-05
Identities = 29/135 (21%), Positives = 67/135 (49%)
Frame = +1
Query: 111 APTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAIS 170
AP +R + +++ K+ ++ +F EM R + T+++L+ + ++ A
Sbjct: 106 APNARSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAWE 285
Query: 171 MFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGW 230
+ G D+ + +L+ LC+ HV+ A L K ++ + D+ T+N++++G
Sbjct: 286 LVDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALV-KKIKDQGIQLDMYTYNILIDGL 462
Query: 231 CVLGNAHEAKRVWKD 245
C G ++A+ ++ D
Sbjct: 463 CKQGRLNDAQVIFHD 507
>TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [imported] -
Arabidopsis thaliana, partial (53%)
Length = 1241
Score = 69.3 bits (168), Expect = 3e-12
Identities = 51/226 (22%), Positives = 105/226 (45%), Gaps = 1/226 (0%)
Frame = +1
Query: 147 VNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETL 206
+ EDT +RR A A + + ++ + D+ + F L+ L ++
Sbjct: 163 IYEDT----VRRLAGAKRFRWVRDIIEHQKSYA-DISNEGFSARLITLYGKSNMHRHAQK 327
Query: 207 FHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMAS-KCRPDLFTYATFIKAL 265
+ + R + + N +L + +R++K + +PDL +Y T+IKAL
Sbjct: 328 LFDEMPQRNCERSVLSLNALLAAYLHSKQYDVVERLFKKLPVQLSVKPDLVSYNTYIKAL 507
Query: 266 TKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEP 325
+KG +A+ + M +G + D++ N ++D L K R + ++++ + E+ P
Sbjct: 508 LEKGSFDSAVSVLEEMEKDGV--ESDLITFNTLLDGLYSKGRFEDGEKLWEKLGEKNVVP 681
Query: 326 NVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLL 371
N+ TYN+ + L +R + E E+ME+K P+ +++ L+
Sbjct: 682 NIRTYNARLLGLAVAKRAGEAVEFYEEMEKK--GVKPDLFSFNALI 813
Score = 58.2 bits (139), Expect = 7e-09
Identities = 45/159 (28%), Positives = 81/159 (50%), Gaps = 7/159 (4%)
Frame = +1
Query: 311 ALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERK---KGSCMPNAVTY 367
A ++F +M +R CE +V + N+L+ K Y++VE + +K + S P+ V+Y
Sbjct: 319 AQKLFDEMPQRNCERSVLSLNALLAAYLH----SKQYDVVERLFKKLPVQLSVKPDLVSY 486
Query: 368 SCLLNSL--KGP-EEVPGVLERMERNGCSLSDDIYNLVLR-LYMKWDNQDGLRKTWDEME 423
+ + +L KG + VLE ME++G +N +L LY K +DG K W+++
Sbjct: 487 NTYIKALLEKGSFDSAVSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDG-EKLWEKLG 663
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
P+ R+Y + G + +A+ ++ EM KG+
Sbjct: 664 EKNVVPNIRTYNARLLGLAVAKRAGEAVEFYEEMEKKGV 780
Score = 55.1 bits (131), Expect = 6e-08
Identities = 36/124 (29%), Positives = 60/124 (48%)
Frame = +1
Query: 151 TFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSK 210
+++T ++ + A+S+ E+ G++ DL F TLL L ED E L+ K
Sbjct: 481 SYNTYIKALLEKGSFDSAVSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDGEKLWE-K 657
Query: 211 AREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGK 270
E + +I+T+N L G V A EA ++++ +PDLF++ IK +G
Sbjct: 658 LGEKNVVPNIRTYNARLLGLAVAKRAGEAVEFYEEMEKKGVKPDLFSFNALIKGFANEGN 837
Query: 271 LGTA 274
L A
Sbjct: 838 LDEA 849
>TC82997 homologue to GP|7293564|gb|AAF48937.1| CG7884 gene product
{Drosophila melanogaster}, partial (2%)
Length = 792
Score = 69.3 bits (168), Expect = 3e-12
Identities = 46/179 (25%), Positives = 82/179 (45%), Gaps = 4/179 (2%)
Frame = +2
Query: 67 VERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKA---DSYAPTSRVCNEILDI 123
+ER L+ + +LV VLR S +L FFNWA Y PTS EI+ I
Sbjct: 236 LERTLNKLRITITPELVFRVLRACSSSPIESLRFFNWAQSLHHHPPYTPTSVEFEEIVTI 415
Query: 124 LGKMSRFEELHQVFDEMSHREGL-VNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDL 182
L + ++ + + +M+H L ++ S+L+ + +++++ +F + F
Sbjct: 416 LANSNNYQTMWSIIHQMTHHHHLSLSPSAVSSLIESYGRHRHIDQSVQLFNKCKIFNCPQ 595
Query: 183 DLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKR 241
+L + +LL LC K A L R+ + D T+ +++N WC G A++
Sbjct: 596 NLQLYNSLLFALCESKLFHAAYALIRRMLRK-GISPDKHTYALLVNAWCSSGKMRXAQQ 769
Score = 40.0 bits (92), Expect = 0.002
Identities = 21/76 (27%), Positives = 40/76 (52%)
Frame = +2
Query: 298 IIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKK 357
+I++ + + +++++F K C N+ YNSL+ LC+ + Y L+ M RK
Sbjct: 512 LIESYGRHRHIDQSVQLFNKCKIFNCPQNLQLYNSLLFALCESKLFHAAYALIRRMLRK- 688
Query: 358 GSCMPNAVTYSCLLNS 373
P+ TY+ L+N+
Sbjct: 689 -GISPDKHTYALLVNA 733
Score = 33.9 bits (76), Expect = 0.15
Identities = 47/243 (19%), Positives = 91/243 (37%), Gaps = 6/243 (2%)
Frame = +2
Query: 114 SRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFY 173
S CN +L K+ R + H + L T ++ FAA V + +
Sbjct: 53 SHKCNAMLHTFAKLIRTTTTLSKSKLLHHHKTLTTTTTTTSKDEYFAAIQHVANIVRRDF 232
Query: 174 TREQFGLDLDLDAFRTLLMWLCRYKHVEDAETL-FHSKAREFQLHRDIKTWNVILNGWC- 231
E+ L + L+ + R E+L F + A+ H +V
Sbjct: 233 YLERTLNKLRITITPELVFRVLRACSSSPIESLRFFNWAQSLHHHPPYTPTSVEFEEIVT 412
Query: 232 VLGNAHEAKRVWKDIMASKCRPDLF----TYATFIKALTKKGKLGTALKLFRGMWNEGCN 287
+L N++ + +W I L ++ I++ + + +++LF + N
Sbjct: 413 ILANSNNYQTMWSIIHQMTHHHHLSLSPSAVSSLIESYGRHRHIDQSVQLFNKC--KIFN 586
Query: 288 CKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVY 347
C ++ + N ++ ALC K A + + M +G P+ TY L+ C +M
Sbjct: 587 CPQNLQLYNSLLFALCESKLFHAAYALIRRMLRKGISPDKHTYALLVNAWCSSGKMRXAQ 766
Query: 348 ELV 350
+ +
Sbjct: 767 QFL 775
Score = 29.6 bits (65), Expect = 2.8
Identities = 10/32 (31%), Positives = 20/32 (62%)
Frame = +2
Query: 422 MERNGCGPDRRSYTIMIHGHYENGKMKDAMRY 453
M R G PD+ +Y ++++ +GKM+ A ++
Sbjct: 677 MLRKGISPDKHTYALLVNAWCSSGKMRXAQQF 772
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,359,937
Number of Sequences: 36976
Number of extensions: 252493
Number of successful extensions: 1850
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 1508
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1706
length of query: 495
length of database: 9,014,727
effective HSP length: 100
effective length of query: 395
effective length of database: 5,317,127
effective search space: 2100265165
effective search space used: 2100265165
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0195b.3


