

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0192.12
(60 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
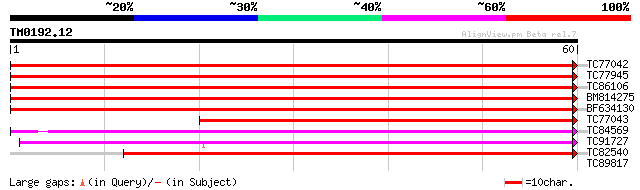
Score E
Sequences producing significant alignments: (bits) Value
TC77042 similar to GP|14532478|gb|AAK63967.1 AT4g11600/T5C23_30 ... 100 2e-22
TC77945 similar to SP|O24296|GSHY_PEA Phospholipid hydroperoxide... 90 2e-19
TC86106 homologue to GP|21068666|emb|CAD31839. putative phosphol... 86 2e-18
BM814275 similar to GP|21592603|gb unknown {Arabidopsis thaliana... 82 4e-17
BF634130 similar to GP|21592603|gb unknown {Arabidopsis thaliana... 80 2e-16
TC77043 similar to PIR|S33618|S33618 glutathione peroxidase (EC ... 67 1e-12
TC84569 weakly similar to GP|727367|gb|AAA64283.1|| Hyr1p {Sacch... 54 2e-08
TC91727 weakly similar to SP|Q9LYB4|GSHZ_ARATH Probable glutathi... 51 1e-07
TC82540 similar to PIR|H83292|H83292 probable glutathione peroxi... 49 4e-07
TC89817 similar to GP|6179604|emb|CAB59895.1 glutathione peroxid... 35 0.004
>TC77042 similar to GP|14532478|gb|AAK63967.1 AT4g11600/T5C23_30
{Arabidopsis thaliana}, partial (74%)
Length = 1015
Score = 99.8 bits (247), Expect = 2e-22
Identities = 49/60 (81%), Positives = 50/60 (82%)
Frame = +1
Query: 1 GFTNSNYTELSPLYEKYK*KGLEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
G TNSNYTELS LYEKYK KGLEILAF CNQF AQEPG E+IQ FV TRFK EFPVFDK
Sbjct: 364 GLTNSNYTELSQLYEKYKSKGLEILAFPCNQFGAQEPGSVEEIQNFVCTRFKAEFPVFDK 543
>TC77945 similar to SP|O24296|GSHY_PEA Phospholipid hydroperoxide
glutathione peroxidase chloroplast precursor (EC
1.11.1.9) (PHGPx)., complete
Length = 1006
Score = 89.7 bits (221), Expect = 2e-19
Identities = 41/60 (68%), Positives = 49/60 (81%)
Frame = +1
Query: 1 GFTNSNYTELSPLYEKYK*KGLEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
G T+SNYTELS LYE +K KGLE+LAF CNQF QEPG +E+I++F TRFK EFP+FDK
Sbjct: 421 GLTSSNYTELSHLYENFKDKGLEVLAFPCNQFGMQEPGSNEEIKKFACTRFKAEFPIFDK 600
>TC86106 homologue to GP|21068666|emb|CAD31839. putative phospholipid
hydroperoxide glutathione peroxidase {Cicer arietinum},
complete
Length = 893
Score = 86.3 bits (212), Expect = 2e-18
Identities = 41/60 (68%), Positives = 46/60 (76%)
Frame = +3
Query: 1 GFTNSNYTELSPLYEKYK*KGLEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
G T +NY EL+ LY+KYK + EILAF CNQFR QEPG SE+IQ V TRFK EFPVFDK
Sbjct: 252 GLTQTNYKELNVLYQKYKDQDFEILAFPCNQFRGQEPGSSEEIQNVVCTRFKAEFPVFDK 431
>BM814275 similar to GP|21592603|gb unknown {Arabidopsis thaliana}, partial
(96%)
Length = 699
Score = 82.0 bits (201), Expect = 4e-17
Identities = 39/60 (65%), Positives = 46/60 (76%)
Frame = +2
Query: 1 GFTNSNYTELSPLYEKYK*KGLEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
G TNSNY L+ LY+KYK KGLEILAF NQF +EPG ++QI +FV T FK EFP+FDK
Sbjct: 176 GMTNSNYVGLNQLYDKYKLKGLEILAFPSNQFGEEEPGTNDQILDFVCTHFKSEFPIFDK 355
>BF634130 similar to GP|21592603|gb unknown {Arabidopsis thaliana}, partial
(90%)
Length = 563
Score = 79.7 bits (195), Expect = 2e-16
Identities = 38/60 (63%), Positives = 45/60 (74%)
Frame = +2
Query: 1 GFTNSNYTELSPLYEKYK*KGLEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
G TNSNY L+ LY+KYK KGLEILAF NQF +EPG ++QI +FV T FK FP+FDK
Sbjct: 155 GMTNSNYVGLNQLYDKYKLKGLEILAFPSNQFGEEEPGTNDQILDFVCTHFKSXFPIFDK 334
>TC77043 similar to PIR|S33618|S33618 glutathione peroxidase (EC 1.11.1.9) -
sweet orange, partial (57%)
Length = 1748
Score = 67.0 bits (162), Expect = 1e-12
Identities = 32/40 (80%), Positives = 33/40 (82%)
Frame = +1
Query: 21 GLEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
GLEILAF CNQF AQEPG E+IQ FV TRFK EFPVFDK
Sbjct: 208 GLEILAFPCNQFGAQEPGSVEEIQNFVCTRFKAEFPVFDK 327
>TC84569 weakly similar to GP|727367|gb|AAA64283.1|| Hyr1p {Saccharomyces
cerevisiae}, partial (79%)
Length = 564
Score = 53.5 bits (127), Expect = 2e-08
Identities = 27/60 (45%), Positives = 34/60 (56%)
Frame = +3
Query: 1 GFTNSNYTELSPLYEKYK*KGLEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
G+T YT L L+ KYK +GL + CNQF AQEPG E I F + + FP+F K
Sbjct: 96 GYT-PQYTGLESLHNKYKDQGLIVAGLPCNQFGAQEPGTEEDIVSFCSVTYNVTFPLFAK 272
>TC91727 weakly similar to SP|Q9LYB4|GSHZ_ARATH Probable glutathione
peroxidase At3g63080 (EC 1.11.1.9). [Mouse-ear cress]
{Arabidopsis thaliana}, partial (57%)
Length = 1124
Score = 50.8 bits (120), Expect = 1e-07
Identities = 31/99 (31%), Positives = 42/99 (42%), Gaps = 40/99 (40%)
Frame = +1
Query: 2 FTNSNYTELSPLYEKYK*----------------------------------------KG 21
F ++NYT+L+ LY KYK G
Sbjct: 133 FADANYTQLTQLYTKYKEIGMHMLFIYLSELMSNIKKILSRILIHVI*FENICLCLLMLG 312
Query: 22 LEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
LEIL F CNQF +EPG S++ Q+F R+K E+P+ K
Sbjct: 313 LEILGFPCNQFLRKEPGTSQEAQDFACDRYKAEYPILGK 429
>TC82540 similar to PIR|H83292|H83292 probable glutathione peroxidase
PA2826 [imported] - Pseudomonas aeruginosa (strain
PAO1), partial (62%)
Length = 678
Score = 48.9 bits (115), Expect = 4e-07
Identities = 21/48 (43%), Positives = 30/48 (61%)
Frame = +2
Query: 13 LYEKYK*KGLEILAFSCNQFRAQEPGDSEQIQEFV*TRFKIEFPVFDK 60
LY KY +GL +L F CNQF QEPG +++I + + FP+F+K
Sbjct: 8 LYTKYAGQGLVVLGFPCNQFGKQEPGGADEISNTCYINYGVSFPMFEK 151
>TC89817 similar to GP|6179604|emb|CAB59895.1 glutathione peroxidase-like
protein GPX54Hv {Hordeum vulgare}, partial (53%)
Length = 584
Score = 35.4 bits (80), Expect = 0.004
Identities = 14/26 (53%), Positives = 20/26 (76%)
Frame = +2
Query: 35 QEPGDSEQIQEFV*TRFKIEFPVFDK 60
QEPG+S + ++F TRFK E+P+F K
Sbjct: 5 QEPGNSLEAEQFACTRFKAEYPIFGK 82
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.337 0.151 0.469
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,556,964
Number of Sequences: 36976
Number of extensions: 12204
Number of successful extensions: 78
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 77
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 78
length of query: 60
length of database: 9,014,727
effective HSP length: 36
effective length of query: 24
effective length of database: 7,683,591
effective search space: 184406184
effective search space used: 184406184
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0192.12


