

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0192.10
(155 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
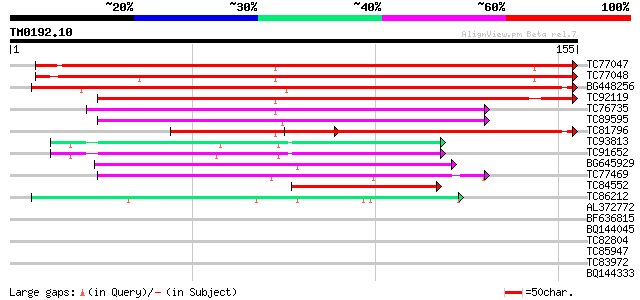
Score E
Sequences producing significant alignments: (bits) Value
TC77047 weakly similar to GP|18252925|gb|AAL62389.1 unknown prot... 216 2e-57
TC77048 similar to GP|18252925|gb|AAL62389.1 unknown protein {Ar... 211 1e-55
BG448256 weakly similar to GP|18252925|gb unknown protein {Arabi... 191 8e-50
TC92119 weakly similar to GP|21739239|emb|CAD40994. OSJNBa0072F1... 135 5e-33
TC76735 similar to GP|5734716|gb|AAD49981.1| Simialr to gb|AF049... 102 6e-23
TC89595 similar to PIR|A96562|A96562 unknown protein [imported] ... 100 3e-22
TC81796 weakly similar to GP|21739239|emb|CAD40994. OSJNBa0072F1... 94 2e-20
TC93813 similar to GP|11935197|gb|AAG42014.1 unknown protein {Ar... 65 8e-12
TC91652 similar to GP|11935197|gb|AAG42014.1 unknown protein {Ar... 61 2e-10
BG645929 similar to GP|22137178|gb AT4g35920/F4B14_190 {Arabidop... 60 4e-10
TC77469 weakly similar to GP|21536856|gb|AAM61188.1 unknown {Ara... 56 5e-09
TC84552 similar to GP|20196932|gb|AAM14839.1 Expressed protein {... 50 4e-07
TC86212 similar to GP|21537192|gb|AAM61533.1 unknown {Arabidopsi... 50 5e-07
AL372772 similar to GP|21928427|dbj seven transmembrane helix re... 33 0.060
BF636815 weakly similar to GP|21593423|gb unknown {Arabidopsis t... 31 0.18
BQ144045 similar to PIR|T51947|T51 probable transcription factor... 31 0.18
TC82804 similar to GP|20385491|gb|AAM21312.1 EMB514 {Arabidopsis... 30 0.30
TC85947 similar to PIR|S52261|S52261 NADH dehydrogenase (ubiquin... 30 0.39
TC83972 similar to PIR|B86414|B86414 hypothetical protein AAF881... 30 0.39
BQ144333 weakly similar to GP|7331719|gb|A Hypothetical protein ... 30 0.39
>TC77047 weakly similar to GP|18252925|gb|AAL62389.1 unknown protein
{Arabidopsis thaliana}, partial (76%)
Length = 873
Score = 216 bits (551), Expect = 2e-57
Identities = 102/157 (64%), Positives = 118/157 (74%), Gaps = 9/157 (5%)
Frame = +2
Query: 8 STDYQPTPPPPPQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGAS 67
STDY P PPPPP+P VEWSTGL DC SD C+T CPC+TFG++AEI+DKGSTSCGAS
Sbjct: 209 STDYAP-PPPPPKPLVEWSTGLCDCCSDPGKSCITLCCPCITFGQVAEIIDKGSTSCGAS 385
Query: 68 GFYF------VQHGVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQ 121
G + + G YS YR+K+R QY LKGN+C DCL HC C CALCQEYRELE +
Sbjct: 386 GALYTLICCVIGCGCLYSCFYRSKMRQQYGLKGNDCTDCLIHCCCEACALCQEYRELENR 565
Query: 122 GFNMKIGWHGNVEQQTRGVAM---TSTAPTVEQSMTR 155
GFNM IGWHGNVEQ+TRG+AM T+TAPTVE M+R
Sbjct: 566 GFNMVIGWHGNVEQRTRGIAMATTTTTAPTVEHGMSR 676
>TC77048 similar to GP|18252925|gb|AAL62389.1 unknown protein {Arabidopsis
thaliana}, partial (73%)
Length = 974
Score = 211 bits (536), Expect = 1e-55
Identities = 100/159 (62%), Positives = 118/159 (73%), Gaps = 11/159 (6%)
Frame = +3
Query: 8 STDYQPTPPPPPQPPVEWSTGLYDCFS---DYNNCCLTYWCPCVTFGRIAEIVDKGSTSC 64
S+DY PPPPP+P VEWSTGL DC S D C+T+WCPC+TFG++AEI+DKGSTSC
Sbjct: 246 SSDY--APPPPPKPLVEWSTGLCDCCSASSDPRKSCITFWCPCITFGQVAEIIDKGSTSC 419
Query: 65 GASGFYF------VQHGVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYREL 118
GASG + + YS YR+K+R QY LKGN+C DCL HC C CALCQEYREL
Sbjct: 420 GASGALYTLICCVIGCPCLYSCFYRSKMRQQYGLKGNDCTDCLIHCCCEACALCQEYREL 599
Query: 119 EKQGFNMKIGWHGNVEQQTRGVAM--TSTAPTVEQSMTR 155
E +GFNM IGWHGNVEQ+TRG+AM T+TAP VEQ M+R
Sbjct: 600 ENRGFNMVIGWHGNVEQRTRGIAMATTTTAPAVEQGMSR 716
>BG448256 weakly similar to GP|18252925|gb unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 649
Score = 191 bits (486), Expect = 8e-50
Identities = 94/166 (56%), Positives = 104/166 (62%), Gaps = 17/166 (10%)
Frame = +2
Query: 7 GSTDYQPTPPPP-----------PQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAE 55
GS QP PP P EWSTGL+DCFSD CC+TYWCPC+TFGRIAE
Sbjct: 128 GSKFQQPYNQPPATGFPVNVGHQPNANQEWSTGLFDCFSDCKTCCITYWCPCITFGRIAE 307
Query: 56 IVDKGSTSCGASGFYFVQH------GVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRC 109
IVDKGSTSC SG + G YS YR K+R QY LK C DCL HC C C
Sbjct: 308 IVDKGSTSCAVSGALYTLICCVTGCGCLYSCIYRNKMRQQYMLKDTPCCDCLVHCCCESC 487
Query: 110 ALCQEYRELEKQGFNMKIGWHGNVEQQTRGVAMTSTAPTVEQSMTR 155
ALCQEYRELE +GF+M++GWHGNV Q +GV M TAP VE MTR
Sbjct: 488 ALCQEYRELENRGFDMELGWHGNVAQGNQGVXMAPTAPAVEH-MTR 622
>TC92119 weakly similar to GP|21739239|emb|CAD40994. OSJNBa0072F16.18 {Oryza
sativa}, partial (60%)
Length = 817
Score = 135 bits (341), Expect = 5e-33
Identities = 67/137 (48%), Positives = 86/137 (61%), Gaps = 6/137 (4%)
Frame = +3
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYF------VQHGVFY 78
WSTGL C D C +T +CPCVTFG IAEIVDKG+++C G + Y
Sbjct: 204 WSTGLCRCLDDPGICLVTCFCPCVTFGLIAEIVDKGNSTCTCDGTIYGALLAVTGLACLY 383
Query: 79 SADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQQTR 138
S YR+K+R+QY+L C+DCL H C CALCQEYREL+ +G+++ IGW NVE+Q
Sbjct: 384 SCYYRSKLRAQYDLPEAPCMDCLVHFCCETCALCQEYRELKNRGYDLSIGWDANVERQRP 563
Query: 139 GVAMTSTAPTVEQSMTR 155
GVA+ AP + MTR
Sbjct: 564 GVAV---APPMISPMTR 605
>TC76735 similar to GP|5734716|gb|AAD49981.1| Simialr to gb|AF049928 PGP224
protein from Petunia x hybrida. {Arabidopsis thaliana},
partial (23%)
Length = 760
Score = 102 bits (254), Expect = 6e-23
Identities = 48/115 (41%), Positives = 64/115 (54%), Gaps = 5/115 (4%)
Frame = +1
Query: 22 PVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYF-----VQHGV 76
P +WS L C + C +T PC+TFG+IAE+VD+G +SC G + +
Sbjct: 91 PPKWSAKLCGCGENPGTCLITCCLPCITFGQIAEVVDEGRSSCAMQGCVYGLLMTITCHW 270
Query: 77 FYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHG 131
YS YR K+R++Y L C DC H C CALCQE+ EL+ +GFN GW G
Sbjct: 271 LYSCLYREKLRAKYGLPAEPCCDCCVHFCCDACALCQEHAELKARGFNPSKGWIG 435
>TC89595 similar to PIR|A96562|A96562 unknown protein [imported] -
Arabidopsis thaliana, partial (63%)
Length = 624
Score = 100 bits (248), Expect = 3e-22
Identities = 46/113 (40%), Positives = 68/113 (59%), Gaps = 6/113 (5%)
Frame = +3
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQ------HGVFY 78
WSTGL+DC + N +T + PCVTFG+IAE++D G SC F ++
Sbjct: 159 WSTGLFDCHENQTNAIMTAFLPCVTFGQIAEVLDGGELSCHLGSFIYLLMMPALCTQWIM 338
Query: 79 SADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHG 131
+ YRTK+R +Y+L D ++H FC C+LCQE+REL+ +G + +GW+G
Sbjct: 339 GSKYRTKLRKKYDLVEAPHTDVISHIFCPCCSLCQEFRELKIRGLDPALGWNG 497
>TC81796 weakly similar to GP|21739239|emb|CAD40994. OSJNBa0072F16.18 {Oryza
sativa}, partial (55%)
Length = 649
Score = 94.4 bits (233), Expect = 2e-20
Identities = 46/80 (57%), Positives = 53/80 (65%)
Frame = +2
Query: 76 VFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQ 135
V+ A TK + LK C DCL HC C CALCQEYRELE +GF+M++GWHGNV Q
Sbjct: 224 VYTHASTVTK*DNNTCLKDTPCCDCLVHCCCESCALCQEYRELENRGFDMELGWHGNVAQ 403
Query: 136 QTRGVAMTSTAPTVEQSMTR 155
+GVAM TAP VE MTR
Sbjct: 404 GNQGVAMAPTAPAVEH-MTR 460
Score = 60.8 bits (146), Expect = 2e-10
Identities = 30/52 (57%), Positives = 33/52 (62%), Gaps = 6/52 (11%)
Frame = +1
Query: 45 CPCVTFGRIAEIVDKGSTSCGASGFYF------VQHGVFYSADYRTKIRSQY 90
CPC+TFGRIAEIVDKGSTSC SG + G YS YR K+R QY
Sbjct: 112 CPCITFGRIAEIVDKGSTSCAVSGALYTLICCVTGCGCLYSCIYRNKMRQQY 267
>TC93813 similar to GP|11935197|gb|AAG42014.1 unknown protein {Arabidopsis
thaliana}, partial (70%)
Length = 664
Score = 65.5 bits (158), Expect = 8e-12
Identities = 42/151 (27%), Positives = 61/151 (39%), Gaps = 43/151 (28%)
Frame = +1
Query: 12 QPTP----PPPPQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEI----------- 56
QP P PP +P W TG++ C D +C +CPCV FGR E
Sbjct: 214 QPLPESYAPPADEP---WMTGIFACVEDRESCLTGLFCPCVLFGRNVESLRENTPWTTPC 384
Query: 57 --------------------------VDKGSTSCGASGFYFV--QHGVFYSADYRTKIRS 88
+D G+T G +F G+ ++ R ++
Sbjct: 385 ICHAIFVEGGISVAIATVIATSFISGIDPGTTCLICEGLFFTWWMCGI-HTGQVRQSLQK 561
Query: 89 QYNLKGNNCLDCLTHCFCSRCALCQEYRELE 119
+Y+LK + C C HC CALCQE+RE++
Sbjct: 562 KYHLKNSPCNACCVHCCLHWCALCQEHREMK 654
>TC91652 similar to GP|11935197|gb|AAG42014.1 unknown protein {Arabidopsis
thaliana}, partial (78%)
Length = 887
Score = 61.2 bits (147), Expect = 2e-10
Identities = 40/148 (27%), Positives = 60/148 (40%), Gaps = 40/148 (27%)
Frame = +1
Query: 12 QPTP----PPPPQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAE------------ 55
QP P PP +P W TG++ C D +C +CPCV FGR E
Sbjct: 130 QPLPESYAPPADEP---WMTGIFGCAEDRESCLTGLFCPCVLFGRNVESLNEDTPWTGPC 300
Query: 56 ----------------------IVDKGSTSCGASGFYFV--QHGVFYSADYRTKIRSQYN 91
++D G++ G +F G+ Y+ R ++ Y+
Sbjct: 301 ICHAIFIEGGIALATATAILNGVIDPGTSFLIFEGLFFTWWMCGI-YTGQVRQNLQKNYH 477
Query: 92 LKGNNCLDCLTHCFCSRCALCQEYRELE 119
L+ + C HC CALCQE+RE++
Sbjct: 478 LQNSPGDPCCVHCCLHWCALCQEHREMK 561
>BG645929 similar to GP|22137178|gb AT4g35920/F4B14_190 {Arabidopsis
thaliana}, partial (24%)
Length = 676
Score = 60.1 bits (144), Expect = 4e-10
Identities = 33/104 (31%), Positives = 50/104 (47%), Gaps = 5/104 (4%)
Frame = -1
Query: 24 EWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF-----Y 78
+W T L C S+ C T++ PC TF +IA + S F+ + + Y
Sbjct: 544 DWHTDLLACCSEPYLCIKTFFYPCGTFSKIATVATNRPISSAEVCNDFIAYSLVLSCCCY 365
Query: 79 SADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQG 122
+ R K+R N+ G D L+H C CAL QE+RE++ +G
Sbjct: 364 TCCIRRKLRKMMNIPGGYVDDFLSHLMCCSCALVQEWREVQIRG 233
>TC77469 weakly similar to GP|21536856|gb|AAM61188.1 unknown {Arabidopsis
thaliana}, partial (53%)
Length = 1150
Score = 56.2 bits (134), Expect = 5e-09
Identities = 39/137 (28%), Positives = 57/137 (41%), Gaps = 30/137 (21%)
Frame = +1
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFY-------------- 70
W + DCF + CPC FG+ + GS A ++
Sbjct: 442 WEGEVLDCFEHRRIALESSCCPCYRFGKNMKRAGLGSCYIQAFVYFLLAICALFNFIAFI 621
Query: 71 -------------FVQHGVFYSADYRTKIRSQYNLKGNNCL--DCLTHCFCSRCALCQEY 115
F+ G Y YRT++R ++N+KG++ + DC+ H C C LCQE
Sbjct: 622 VTRHHYFLYLTVTFIITGGAYLGFYRTRMRKKFNIKGSDSMVDDCVYHFVCPCCTLCQES 801
Query: 116 RELEKQGFNMKIG-WHG 131
R LE N++ G WHG
Sbjct: 802 RTLEIN--NVQDGTWHG 846
>TC84552 similar to GP|20196932|gb|AAM14839.1 Expressed protein {Arabidopsis
thaliana}, partial (30%)
Length = 510
Score = 50.1 bits (118), Expect = 4e-07
Identities = 21/41 (51%), Positives = 27/41 (65%)
Frame = +3
Query: 78 YSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYREL 118
Y+ YR +RS+YNL C D +TH C CA+CQEYRE+
Sbjct: 81 YACGYRNTLRSKYNLPEAPCGDFVTHFCCHLCAICQEYREI 203
>TC86212 similar to GP|21537192|gb|AAM61533.1 unknown {Arabidopsis
thaliana}, partial (78%)
Length = 1237
Score = 49.7 bits (117), Expect = 5e-07
Identities = 49/162 (30%), Positives = 62/162 (38%), Gaps = 44/162 (27%)
Frame = +2
Query: 7 GSTDYQPTPPPPPQPPVEWSTGLYD--CFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSC 64
GS QP P P G D C SD C L PCV +G E + +
Sbjct: 326 GSVMGQPIPRSPWNSSACACLGQNDHFCSSDLEVCLLGSVAPCVLYGSNMERLHSNPGTF 505
Query: 65 GA-----SGFYFVQHGVF--------YSADYRTKIRSQYNLKGN------NC-------- 97
G SG Y + + F S RT+IR ++NL+G+ +C
Sbjct: 506 GNHCLHYSGLYVIGNSCFGWNCLAPWLSYHSRTEIRRRFNLEGSCEALNRSCGCCGSYLE 685
Query: 98 -----------LDCLTHCFCSRCALCQEYRELEKQ----GFN 124
D TH FC CALCQE REL ++ GFN
Sbjct: 686 NEEQREHYELACDFATHVFCHVCALCQEGRELRRRVPHPGFN 811
>AL372772 similar to GP|21928427|dbj seven transmembrane helix receptor {Homo
sapiens}, partial (12%)
Length = 384
Score = 32.7 bits (73), Expect = 0.060
Identities = 25/83 (30%), Positives = 36/83 (43%), Gaps = 1/83 (1%)
Frame = +3
Query: 39 CCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVFYSADYRTKIRSQYNLKGNNC- 97
CCL YW C T+GR +C Y+ + R+ + + L C
Sbjct: 15 CCL-YWLLC-TYGRCC-------MAC-------------YAGNMRSALFMKLGLPDPGCC 128
Query: 98 LDCLTHCFCSRCALCQEYRELEK 120
++ L HC C+ CAL QE R +K
Sbjct: 129 INWLCHCCCAPCALSQEGRAAKK 197
>BF636815 weakly similar to GP|21593423|gb unknown {Arabidopsis thaliana},
partial (56%)
Length = 620
Score = 31.2 bits (69), Expect = 0.18
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = -1
Query: 12 QPTPPPPPQPPVEWSTGLYDCFSDYNN 38
QP PPPPP PP S+ L S ++N
Sbjct: 281 QPPPPPPPSPPPPSSSRLLISLSHFSN 201
>BQ144045 similar to PIR|T51947|T51 probable transcription factor HUA2
[imported] - Arabidopsis thaliana, partial (1%)
Length = 1172
Score = 31.2 bits (69), Expect = 0.18
Identities = 11/16 (68%), Positives = 11/16 (68%)
Frame = +3
Query: 13 PTPPPPPQPPVEWSTG 28
P PPPPP PP WS G
Sbjct: 45 PPPPPPPPPPPCWSVG 92
>TC82804 similar to GP|20385491|gb|AAM21312.1 EMB514 {Arabidopsis thaliana},
partial (43%)
Length = 624
Score = 30.4 bits (67), Expect = 0.30
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = +1
Query: 13 PTPPPPPQPPVEWST 27
PTPPPPP PP W +
Sbjct: 88 PTPPPPPPPP*TWKS 132
>TC85947 similar to PIR|S52261|S52261 NADH dehydrogenase (ubiquinone) (EC
1.6.5.3) flavoprotein 1 precursor - potato, partial
(44%)
Length = 1086
Score = 30.0 bits (66), Expect = 0.39
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = +1
Query: 1 MYKTVAGSTDYQPTPPPPPQPP 22
++ T ST P PPPPP PP
Sbjct: 85 LFSTQGASTASTPQPPPPPPPP 150
>TC83972 similar to PIR|B86414|B86414 hypothetical protein AAF88121.1
[imported] - Arabidopsis thaliana, partial (4%)
Length = 1037
Score = 30.0 bits (66), Expect = 0.39
Identities = 12/25 (48%), Positives = 15/25 (60%), Gaps = 2/25 (8%)
Frame = +2
Query: 3 KTVAGSTDYQPT--PPPPPQPPVEW 25
K V + +PT PPPPP PP +W
Sbjct: 434 KDVTSHSHAEPTRPPPPPPVPPTQW 508
>BQ144333 weakly similar to GP|7331719|gb|A Hypothetical protein
Y110A2AL.12 {Caenorhabditis elegans}, partial (8%)
Length = 1163
Score = 30.0 bits (66), Expect = 0.39
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = +1
Query: 13 PTPPPPPQPPVEWS 26
PTPPPPP P + WS
Sbjct: 31 PTPPPPPVPRLRWS 72
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,590,254
Number of Sequences: 36976
Number of extensions: 166894
Number of successful extensions: 5267
Number of sequences better than 10.0: 199
Number of HSP's better than 10.0 without gapping: 2351
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4384
length of query: 155
length of database: 9,014,727
effective HSP length: 88
effective length of query: 67
effective length of database: 5,760,839
effective search space: 385976213
effective search space used: 385976213
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0192.10


