

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
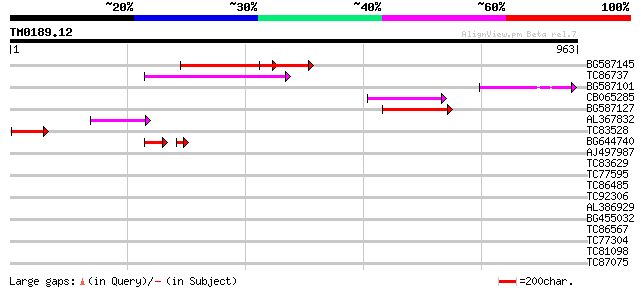
Query= TM0189.12
(963 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 158 8e-39
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 134 2e-31
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 96 5e-20
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 90 5e-18
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 89 1e-17
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 77 3e-14
TC83528 62 1e-09
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 44 1e-05
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 39 0.007
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 37 0.048
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 35 0.14
TC86485 similar to GP|23315211|gb|AAN20392.1 Sequence 405 from p... 33 0.53
TC92306 32 1.5
AL386929 30 3.4
BG455032 weakly similar to GP|10440622|gb| putative NBS-LRR type... 30 3.4
TC86567 similar to GP|15384989|emb|CAC59823. Xaa-Pro aminopeptid... 30 5.8
TC77304 similar to GP|10177347|dbj|BAB10690. peptidylprolyl isom... 30 5.8
TC81098 homologue to GP|20373023|dbj|BAB91180. lipoic acid synth... 29 7.6
TC87075 similar to GP|16209674|gb|AAL14395.1 AT5g15050/F2G14_170... 29 9.9
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 158 bits (400), Expect = 8e-39
Identities = 75/165 (45%), Positives = 108/165 (65%)
Frame = +2
Query: 290 MHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVK 349
MH D +KT F+T R YCY+ MPFGLKNAG+TYQRL++R+F ++G MEVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 350 SVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQ 409
S+ ++H L E F + ++ M+LNP KC+FG+ G+FLG+++T + IE+NP + AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 410 EMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTF 454
++ SP N +EVQRL G L PF++ + TF
Sbjct: 362 DLPSPKNSREVQRLTGEDRGAEPLHLQVHR*MPPFLQATLRKQTF 496
Score = 94.4 bits (233), Expect = 2e-19
Identities = 46/92 (50%), Positives = 62/92 (67%)
Frame = +3
Query: 425 GRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQGL 484
GRI AL+RF+ S DK PF K L N F W+ +CEEAF +LK+ L+ PPVLSKP G
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGD 587
Query: 485 PLHLYFSVGDHAISSVILQEVDGEQKIVYFVS 516
L LY ++ A+SSV+++E GEQK +++ S
Sbjct: 588 TLSLYIAISSTAVSSVLIREDRGEQKPIFYTS 683
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 134 bits (336), Expect = 2e-31
Identities = 72/250 (28%), Positives = 128/250 (50%), Gaps = 2/250 (0%)
Frame = +1
Query: 230 VVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIK 289
V+ V+K G R CVD LN KD YPLP I + + G + +D + +H+++
Sbjct: 583 VLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMR 762
Query: 290 MHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVK 349
+ D++KT F T + + PFGL A AT+QR +++ + + Y+DD+++
Sbjct: 763 IKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIY 942
Query: 350 SVLGSNHHE-DLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITS-RRIEINPDKCKA 407
+ HE + R+ + L+P+KC F + K++GF++T+ + + +P K A
Sbjct: 943 TTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAA 1122
Query: 408 IQEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRL 467
I++ P +VK + +G F+ + + P + +K+ F W +E E AFT+L
Sbjct: 1123IRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFTKL 1302
Query: 468 KEMLSAPPVL 477
K + + PVL
Sbjct: 1303KRLFAEEPVL 1332
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 96.3 bits (238), Expect = 5e-20
Identities = 52/167 (31%), Positives = 87/167 (51%), Gaps = 3/167 (1%)
Frame = +2
Query: 799 REAGHYTLLDGILFRRGFSSPLLRCLPPEKYEAVMTEVHEGVCASHIGGRSLASKVLRAG 858
++ Y + L+++ + +RC+ E+ ++ H A H SK+ +AG
Sbjct: 5 KDVRRYLWDEPFLYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAG 184
Query: 859 FYWPTIRKECAEFVRKCKKCQVFTDLP---RAPPGQLVTISSPWPFAMWGVDLVGPFPTA 915
F+WPT+ K+ F+ KC CQ ++ P ++ + F +WG+D +GPFP++
Sbjct: 185 FWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMPQNFILEVE---VFDVWGIDFMGPFPSS 355
Query: 916 RSQMKFILVAVDYFTKWVEAEPLASITAAKIISFYWKRIVCRFGVPR 962
+ K+ILVAVDY +KWVEA + A ++ + I RFGVPR
Sbjct: 356 YNN-KYILVAVDYVSKWVEAIASPTNDATVVVKMFKSVIFPRFGVPR 493
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 89.7 bits (221), Expect = 5e-18
Identities = 50/133 (37%), Positives = 75/133 (55%)
Frame = -2
Query: 609 EGEKVSTQWILFVDGSSNDNGSEAGVTLQGPGELVLEQSLRFQFKTSNNQAEYEALIAGL 668
EG +++W L DG+ N G G + P + + R F+ +NN AEYEA I G+
Sbjct: 558 EGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGI 379
Query: 669 KLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQQVVVEFVPRA 728
+ AI+++I L + DS LV NQ+ GE++ LI Y + R L+ +V + +PR
Sbjct: 378 EEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHHIPRD 199
Query: 729 QNQRADALAKLAS 741
+NQ ADALA L+S
Sbjct: 198 ENQMADALATLSS 160
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 88.6 bits (218), Expect = 1e-17
Identities = 46/119 (38%), Positives = 75/119 (62%)
Frame = +3
Query: 633 GVTLQGPGELVLEQSLRFQFKTSNNQAEYEALIAGLKLAIEVQIDSLPVRTDSPLVANQV 692
G+ L P +L+QS R +F SNN+ YEALIAG++LA ++I ++ DS LVA+Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 693 NGEFQVKELALIKYVERVRLLMGRLQQVVVEFVPRAQNQRADALAKLASTRKPGNNRSV 751
+GE++ ++ + Y++ V+ L +L + +PR++N +ADALA LAS+ P R +
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALASSSDPELKRVI 359
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 77.0 bits (188), Expect = 3e-14
Identities = 37/102 (36%), Positives = 60/102 (58%)
Frame = -1
Query: 137 LKIGTKLSEEQEQRISKLLGDNLDLFAWSHKYMPGIDPNFICHRLALNPGVEPITQTRRR 196
+K+G L E +++I +LL + LD+FA S++ MPG+DP + HR+ P P+ RR
Sbjct: 306 IKVGAALEEGVKRKIFQLLREYLDIFACSYEDMPGLDPKIVEHRIPTKPECPPVR*KLRR 127
Query: 197 MGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANG 238
+ I+ EV K + A F+ ++YP W+ N+V V K +G
Sbjct: 126 THPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDG 1
>TC83528
Length = 555
Score = 61.6 bits (148), Expect = 1e-09
Identities = 29/66 (43%), Positives = 46/66 (68%), Gaps = 3/66 (4%)
Frame = +3
Query: 4 VKDALDLDTTFGEEENARTLSVKYLILETEGS---YNMIIGRITLNRLCAVISTAQLAVK 60
VK +DLDTTFG+ EN +T+ V+Y ++E+ S YN+++G +L L AV+S A+ +K
Sbjct: 357 VKGYIDLDTTFGKGENTKTIKVRYFVVESPPSVSIYNVVLGWPSLKDLKAVLSVAEFTIK 536
Query: 61 YPLSNG 66
YP+ +G
Sbjct: 537 YPVGDG 554
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 43.5 bits (101), Expect(2) = 1e-05
Identities = 18/39 (46%), Positives = 25/39 (63%)
Frame = -1
Query: 230 VVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVD 268
++ V+K +G +RMC+D NK K+ YPLP ID L D
Sbjct: 232 LLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFD 116
Score = 24.3 bits (51), Expect(2) = 1e-05
Identities = 9/21 (42%), Positives = 15/21 (70%)
Frame = -3
Query: 284 GYHQIKMHQSDEDKTTFMTAR 304
GYHQ ++++ + KTT +T R
Sbjct: 71 GYHQFRVNEVNIPKTTLLTWR 9
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 39.3 bits (90), Expect = 0.007
Identities = 22/60 (36%), Positives = 30/60 (49%)
Frame = -2
Query: 609 EGEKVSTQWILFVDGSSNDNGSEAGVTLQGPGELVLEQSLRFQFKTSNNQAEYEALIAGL 668
EG + W L DG+ N G+ G L P + + R +F +NN AEYEA I G+
Sbjct: 314 EGPDPDSVWGLIFDGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 36.6 bits (83), Expect = 0.048
Identities = 35/114 (30%), Positives = 49/114 (42%), Gaps = 5/114 (4%)
Frame = +2
Query: 625 SNDNGSEAG---VTLQGPGELV--LEQSLRFQFKTSNNQAEYEALIAGLKLAIEVQIDSL 679
SN N AG + L G L+ Q L Q K S AEY AL+ GLK A +
Sbjct: 578 SNINRGPAGAGALLLAEDGSLLYGFRQGLGHQTKES---AEYRALLLGLKHASMKGFKYV 748
Query: 680 PVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQQVVVEFVPRAQNQRA 733
+ DS LV NQ+ +++K+ L K L ++ + R +N A
Sbjct: 749 TAKGDSELVINQILDPWKIKDEHLKKLCAEALELSDNFHSFRIQHISRERNYGA 910
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 35.0 bits (79), Expect = 0.14
Identities = 28/114 (24%), Positives = 50/114 (43%), Gaps = 2/114 (1%)
Frame = +2
Query: 832 VMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQ 891
++ E H+ A H GR+ +++ F+WP + FVR C C +A G
Sbjct: 179 LVQESHDSTAAGH-PGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGF 355
Query: 892 LVTISSPWPF-AMWGVDLVGPFPTARSQ-MKFILVAVDYFTKWVEAEPLASITA 943
L + P + +D + P R + +++ V VD +K V E + ++ A
Sbjct: 356 LKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDTMEA 517
>TC86485 similar to GP|23315211|gb|AAN20392.1 Sequence 405 from patent US
6410718, partial (90%)
Length = 1927
Score = 33.1 bits (74), Expect = 0.53
Identities = 15/42 (35%), Positives = 25/42 (58%)
Frame = +3
Query: 411 MKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNT 452
MKSP ++K+VQ+ + + LSR + S + ++KC K T
Sbjct: 1131 MKSPEDLKKVQQELAEVVGLSRQVEESDFEKLTYLKCALKET 1256
>TC92306
Length = 521
Score = 31.6 bits (70), Expect = 1.5
Identities = 24/73 (32%), Positives = 29/73 (38%)
Frame = +2
Query: 665 IAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQQVVVEF 724
I GL A + + VR D V Q G ++V L L + V VE
Sbjct: 2 ILGLNEARNQGYEHVHVRGDFQXVCKQFXGSWKVNNPNLRNLCNXAVELKSNFKSVSVEH 181
Query: 725 VPRAQNQRADALA 737
VPR N ADA A
Sbjct: 182 VPRGXNXAADAQA 220
>AL386929
Length = 474
Score = 30.4 bits (67), Expect = 3.4
Identities = 18/49 (36%), Positives = 25/49 (50%)
Frame = +1
Query: 119 NRPTPIEETKDLKFGEKILKIGTKLSEEQEQRISKLLGDNLDLFAWSHK 167
N P PIEE K G++ +K K EE E + K L +N + W+ K
Sbjct: 85 NLPLPIEENKLRILGKESIK---KFKEEVESKERKALTENEKIEGWNKK 222
>BG455032 weakly similar to GP|10440622|gb| putative NBS-LRR type resistance
protein 3' partial {Oryza sativa}, partial (3%)
Length = 666
Score = 30.4 bits (67), Expect = 3.4
Identities = 19/64 (29%), Positives = 32/64 (49%), Gaps = 1/64 (1%)
Frame = +1
Query: 232 IVKKANGKWRMCVDNTDLNKTCPKDSYP-LPSIDKLVDGALGNELLSLMDAYSGYHQIKM 290
++K G+ C+ + D+ K CPK P LPS+ L NELL + + G Q+ +
Sbjct: 64 LLKVERGEMFPCLSSLDIWK-CPKLGLPCLPSLKDLGVDGRNNELLRSISTFRGLTQLTL 240
Query: 291 HQSD 294
+ +
Sbjct: 241 NSGE 252
>TC86567 similar to GP|15384989|emb|CAC59823. Xaa-Pro aminopeptidase 1
{Lycopersicon esculentum}, partial (91%)
Length = 2223
Score = 29.6 bits (65), Expect = 5.8
Identities = 14/36 (38%), Positives = 20/36 (54%)
Frame = -2
Query: 360 LMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITS 395
++ AFG ++ +LNP KCSF R L +TS
Sbjct: 1343 IIAAFGPTEEIVGKLNPLKCSFDARNSSSLSLTVTS 1236
>TC77304 similar to GP|10177347|dbj|BAB10690. peptidylprolyl isomerase
{Arabidopsis thaliana}, partial (74%)
Length = 1627
Score = 29.6 bits (65), Expect = 5.8
Identities = 14/36 (38%), Positives = 23/36 (63%)
Frame = +3
Query: 244 VDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLM 279
+D+ D+NK+ PKD P+ SID + N L+S++
Sbjct: 1359 LDSLDINKSAPKDVEPM-SIDSKA*STMTNSLISVL 1463
>TC81098 homologue to GP|20373023|dbj|BAB91180. lipoic acid synthase
{Arabidopsis thaliana}, partial (44%)
Length = 842
Score = 29.3 bits (64), Expect = 7.6
Identities = 13/37 (35%), Positives = 19/37 (51%)
Frame = +3
Query: 873 RKCKKCQVFTDLPRAPPGQLVTISSPWPFAMWGVDLV 909
R C+ C V T + APP + ++ A WGVD +
Sbjct: 522 RGCRFCAVKTSINPAPPDPMEPENTAKAIASWGVDYI 632
>TC87075 similar to GP|16209674|gb|AAL14395.1 AT5g15050/F2G14_170
{Arabidopsis thaliana}, partial (67%)
Length = 1863
Score = 28.9 bits (63), Expect = 9.9
Identities = 22/61 (36%), Positives = 27/61 (44%)
Frame = +1
Query: 429 ALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQGLPLHL 488
+LSRFLLHS S P L + + EE +LS P L P PLHL
Sbjct: 52 SLSRFLLHSLFSSTPKPLSLSLANGNQQTTTQEEKMVPTTHLLSTPHHLPHP----PLHL 219
Query: 489 Y 489
+
Sbjct: 220 H 222
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,340,590
Number of Sequences: 36976
Number of extensions: 401235
Number of successful extensions: 1711
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1647
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1706
length of query: 963
length of database: 9,014,727
effective HSP length: 105
effective length of query: 858
effective length of database: 5,132,247
effective search space: 4403467926
effective search space used: 4403467926
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0189.12


